Nie możesz wybrać więcej, niż 25 tematów
Tematy muszą się zaczynać od litery lub cyfry, mogą zawierać myślniki ('-') i mogą mieć do 35 znaków.
1.1KB
1.1KB
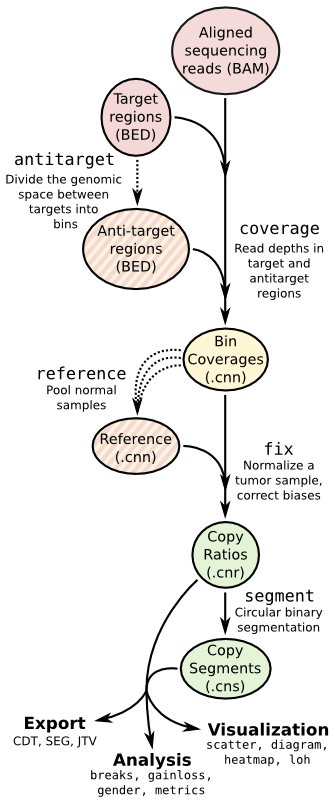
CNVkit
Author: Yaqing Liu
E-mail:yaqing.liu@outlook.com
Install
# activate choppy environment
open-choppy-env
# install app
choppy install YaqingLiu/CNVkit
Copy number calling pipeline
Input
{
"tumor_bam": [
"oss://choppy-cromwell-result/...bam",
"oss://choppy-cromwell-result/...bam",
"oss://choppy-cromwell-result/...bam"
],
"tumor_bai": [
"oss://choppy-cromwell-result/...bai",
"oss://choppy-cromwell-result/...bai",
"oss://choppy-cromwell-result/...bai"
],
"normal_bam": [
"oss://choppy-cromwell-result/...bam",
"oss://choppy-cromwell-result/...bam",
"oss://choppy-cromwell-result/...bam"
],
"normal_bai": [
"oss://choppy-cromwell-result/...bai",
"oss://choppy-cromwell-result/...bai",
"oss://choppy-cromwell-result/...bai"
],
"sample_id": "..."
}
Output
A segment file and some intermediate *.cnn/cns will be generated, and the segment file can be imported into IGV.