You can not select more than 25 topics
Topics must start with a letter or number, can include dashes ('-') and can be up to 35 characters long.
|
|
преди 4 години | |
|---|---|---|
| assest | преди 4 години | |
| tasks | преди 4 години | |
| README.md | преди 4 години | |
| defaults | преди 4 години | |
| inputs | преди 4 години | |
| workflow.wdl | преди 4 години | |
README.md
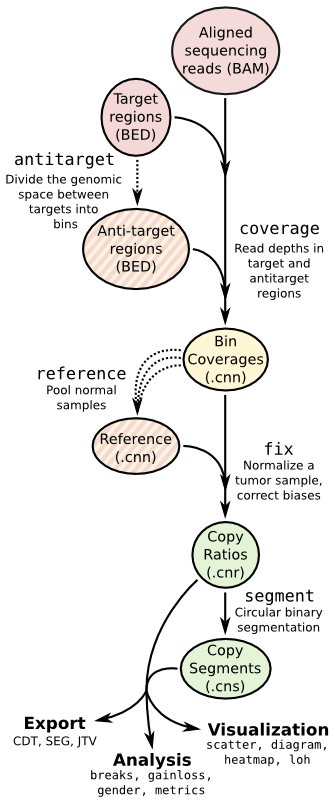
CNVkit
Author: Yaqing Liu
E-mail: yaqing.liu@outlook.com
CNVkit is a Python library and command-line software toolkit to infer and visualize copy number from high-throughput DNA sequencing data. It is designed for use with hybrid capture, including both whole-exome and custom target panels, and short-read sequencing platforms such as Illumina and Ion Torrent.
Official document: https://cnvkit.readthedocs.io/en/stable/index.html
Install
# activate choppy environment
open-choppy-env
# install app
choppy install YaqingLiu/CNVkit
Copy number calling pipeline
Input
{
"tumor_bam": [
"oss://choppy-cromwell-result/...bam",
"oss://choppy-cromwell-result/...bam",
"oss://choppy-cromwell-result/...bam"
],
"tumor_bai": [
"oss://choppy-cromwell-result/...bai",
"oss://choppy-cromwell-result/...bai",
"oss://choppy-cromwell-result/...bai"
],
"normal_bam": [
"oss://choppy-cromwell-result/...bam",
"oss://choppy-cromwell-result/...bam",
"oss://choppy-cromwell-result/...bam"
],
"normal_bai": [
"oss://choppy-cromwell-result/...bai",
"oss://choppy-cromwell-result/...bai",
"oss://choppy-cromwell-result/...bai"
],
"sample_id": "..."
}
Output
A segment file and some intermediate *.cnn/cns will be generated, and the segment file can be imported into IGV.