Nie możesz wybrać więcej, niż 25 tematów
Tematy muszą się zaczynać od litery lub cyfry, mogą zawierać myślniki ('-') i mogą mieć do 35 znaków.
|
|
4 lat temu | |
|---|---|---|
| assest | 4 lat temu | |
| tasks | 4 lat temu | |
| README.md | 4 lat temu | |
| defaults | 4 lat temu | |
| inputs | 4 lat temu | |
| workflow.wdl | 4 lat temu | |
README.md
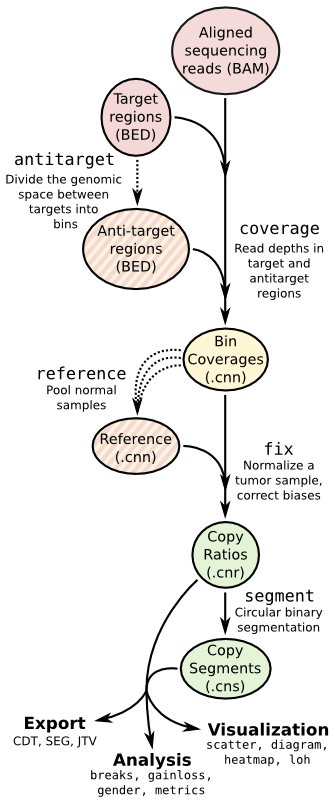
CNVkit
Author: Yaqing Liu
E-mail: yaqing.liu@outlook.com
CNVkit is a Python library and command-line software toolkit to infer and visualize copy number from high-throughput DNA sequencing data. It is designed for use with hybrid capture, including both whole-exome and custom target panels, and short-read sequencing platforms such as Illumina and Ion Torrent.
Official document: https://cnvkit.readthedocs.io/en/stable/index.html
Install
# activate choppy environment
open-choppy-env
# install app
choppy install YaqingLiu/CNVkit
Copy number calling pipeline
Input
{
"tumor_bam": [
"oss://choppy-cromwell-result/...bam",
"oss://choppy-cromwell-result/...bam",
"oss://choppy-cromwell-result/...bam"
],
"tumor_bai": [
"oss://choppy-cromwell-result/...bai",
"oss://choppy-cromwell-result/...bai",
"oss://choppy-cromwell-result/...bai"
],
"normal_bam": [
"oss://choppy-cromwell-result/...bam",
"oss://choppy-cromwell-result/...bam",
"oss://choppy-cromwell-result/...bam"
],
"normal_bai": [
"oss://choppy-cromwell-result/...bai",
"oss://choppy-cromwell-result/...bai",
"oss://choppy-cromwell-result/...bai"
],
"sample_id": "...",
"method": "...",
"reference": "..." # this parameter is optional
}
Note
-m {hybrid,amplicon,wgs}, --seq-method {hybrid,amplicon,wgs}, --method {hybrid,amplicon,wgs}
Sequencing assay type: hybridization capture
('hybrid'), targeted amplicon sequencing
('amplicon'), or whole genome sequencing ('wgs').
Determines whether and how to use antitarget bins.
To reuse an existing reference or create a new :
-r REFERENCE, --reference REFERENCE
Copy number reference file (.cnn).
--output-reference FILENAME
Output filename/path for the new reference file
being created. (If given, ignores the
-o/--output-dir option and will write the file to
the given path. Otherwise, "reference.cnn" will
be created in the current directory or specified
output directory.)
Output
- *.cnn/cns of each sample.
- A whole-genome copy ratio profile as a PDF scatter plot.
- An ideogram of copy ratios on chromosomes as a PDF.
- A segment file which can be imported into IGV.