2.7KB
CNVkit
Author: Yaqing Liu
E-mail: yaqing.liu@outlook.com
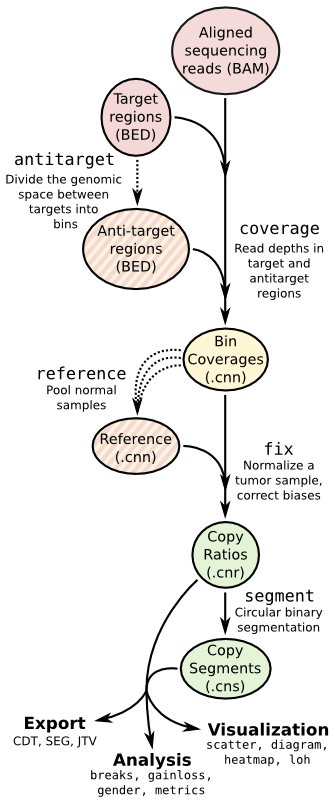
CNVkit is a Python library and command-line software toolkit to infer and visualize copy number from high-throughput DNA sequencing data. It is designed for use with hybrid capture, including both whole-exome and custom target panels, and short-read sequencing platforms such as Illumina and Ion Torrent.
Official document: https://cnvkit.readthedocs.io/en/stable/index.html
Install
# activate choppy environment
open-choppy-env
# install app
choppy install YaqingLiu/CNVkit
Copy number calling pipeline
Input
{
"tumor_bam": [
"oss://choppy-cromwell-result/...bam",
"oss://choppy-cromwell-result/...bam",
"oss://choppy-cromwell-result/...bam"
],
"tumor_bai": [
"oss://choppy-cromwell-result/...bai",
"oss://choppy-cromwell-result/...bai",
"oss://choppy-cromwell-result/...bai"
],
"normal_bam": [
"oss://choppy-cromwell-result/...bam",
"oss://choppy-cromwell-result/...bam",
"oss://choppy-cromwell-result/...bam"
],
"normal_bai": [
"oss://choppy-cromwell-result/...bai",
"oss://choppy-cromwell-result/...bai",
"oss://choppy-cromwell-result/...bai"
],
"sample_id": "...",
"method": "...",
"reference": "..." # this parameter is optional
}
Note -m {hybrid,amplicon,wgs}, --seq-method {hybrid,amplicon,wgs}, --method {hybrid,amplicon,wgs}
Sequencing assay type: hybridization capture ('hybrid'), targeted amplicon sequencing ('amplicon'), or whole genome sequencing ('wgs').
Determines whether and how to use antitarget bins.
To reuse an existing reference or create a new : -r REFERENCE, --reference REFERENCE
Copy number reference file (.cnn).
--output-reference FILENAME
Output filename/path for the new reference file being created. (If given, ignores the -o/--output-dir option and will write the file to the given path. Otherwise, "reference.cnn" will be created in the current directory or specified output directory.)
--annotate The gene annotations file (refFlat.txt) is useful to apply gene names to your baits BED file, if the BED file does not already have short, informative names for each bait interval. This file can be used in the next step. If the BED looks like this:
chr1 1508981 1509154 SSU72 chr1 2407978 2408183 PLCH2 chr1 2409866 2410095 PLCH2 Then you don’t need refFlat.txt.
Output
- *.cnn/cns of each sample.
- A whole-genome copy ratio profile as a PDF scatter plot.
- An ideogram of copy ratios on chromosomes as a PDF.
- A segment file which can be imported into IGV.