共有 3 个文件被更改,包括 10 次插入 和 12 次删除
+ 2
- 0
README.md
查看文件
| @@ -31,6 +31,8 @@ choppy install renluyao/WGS_germline_datapotal | |||
|  | |||
|  | |||
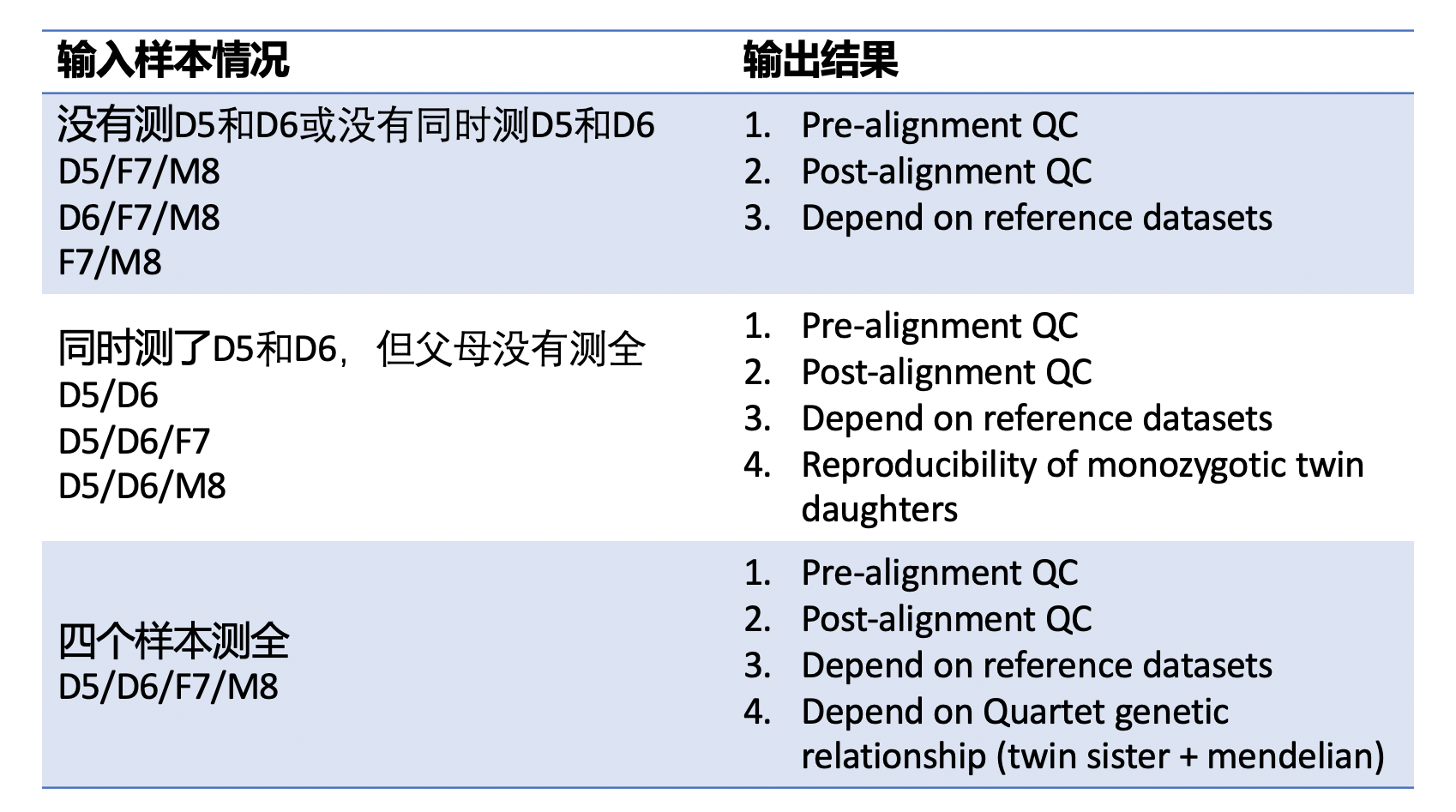
| ### 1. 原始数据质量控制 | |||
| #### [Fastqc](<https://www.bioinformatics.babraham.ac.uk/projects/fastqc/>) v0.11.5 | |||
+ 8
- 12
codescripts/D5_D6.py
查看文件
| @@ -29,14 +29,14 @@ snv_sister_diff = 0 | |||
| for row in sister_dat.itertuples(): | |||
| # snv indel | |||
| if ',' in row.ALT: | |||
| alt = row.ALT.split(',') | |||
| if ',' in row[4]: | |||
| alt = row[4].split(',') | |||
| alt_len = [len(i) for i in alt] | |||
| alt_max = max(alt_len) | |||
| else: | |||
| alt_max = len(row.ALT) | |||
| alt_max = len(row[4]) | |||
| alt = alt_max | |||
| ref = row.REF | |||
| ref = row[3] | |||
| if len(ref) == 1 and alt == 1: | |||
| cate = 'SNV' | |||
| elif len(ref) > alt: | |||
| @@ -62,12 +62,6 @@ for row in sister_dat.itertuples(): | |||
| sister_count = "no" | |||
| else: | |||
| sister_count = "yes_diff" | |||
| if sister_count == 'yes_same': | |||
| sister_same += 1 | |||
| elif sister_count == 'yes_diff': | |||
| sister_diff += 1 | |||
| else: | |||
| pass | |||
| if cate == 'SNV': | |||
| if sister_count == 'yes_same': | |||
| snv_sister_same += 1 | |||
| @@ -86,9 +80,11 @@ for row in sister_dat.itertuples(): | |||
| indel_sister = indel_sister_same/(indel_sister_same + indel_sister_diff) | |||
| snv_sister = snv_sister_same/(snv_sister_same + snv_sister_diff) | |||
| outcolumn = 'Project\tReproducibility_D5_D6\n' | |||
| outResult = project_name + '\t' + str(sister) + '\n' | |||
| indel_outResult = project_name + '.INDEL' + '\t' + str(indel_sister) + '\n' | |||
| snv_outResult = project_name + '.SNV' + '\t' + str(snv_sister) + '\n' | |||
| output_file.write(outcolumn) | |||
| output_file.write(outResult) | |||
| output_file.write(indel_outResult) | |||
| output_file.write(snv_outResult) | |||
二进制
pictures/table.png
查看文件
正在加载...