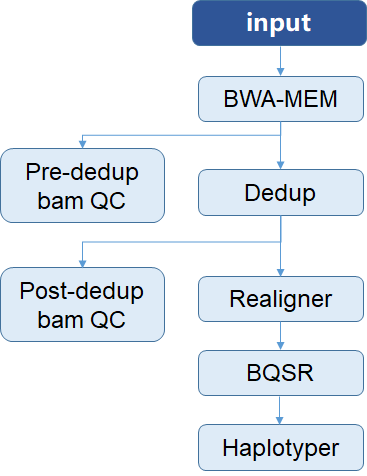
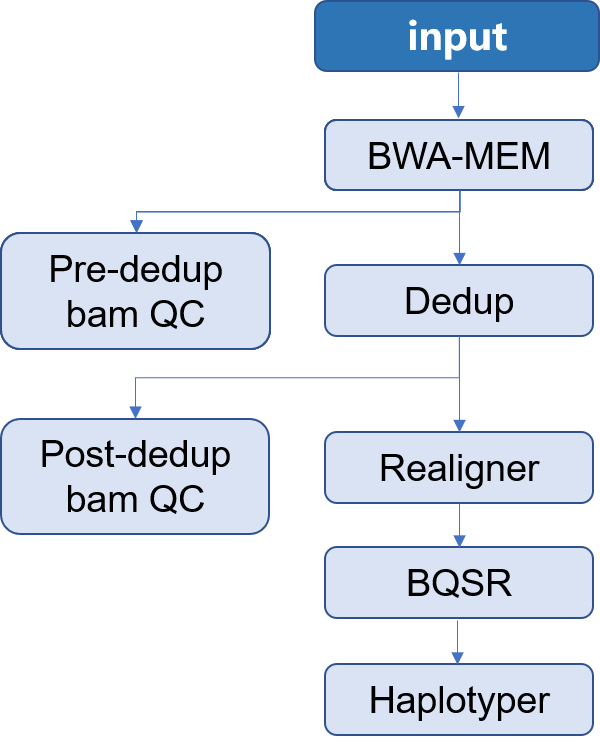
19 arquivos alterados com 354 adições e 473 exclusões
BIN
.DS_Store
Ver arquivo
+ 0
- 0
LICENSE.md
Ver arquivo
+ 53
- 0
README.md
Ver arquivo
| @@ -0,0 +1,53 @@ | |||
| ### Variant Calling | |||
| This APP developed for somatic short variant discovery (SNVs + Indels). | |||
| ***Supported callers*** | |||
| * TNseq (TNhaplotyper2) | |||
| * TNscope | |||
| * VarScan | |||
| * TNhaplotyper (This caller is only available in `v0.1.0` as it is too outdated) | |||
| Variant caller can be selected by setting `ture/false` in the submitted sample.csv. | |||
| ***Accepted data*** | |||
| * TN matched WES | |||
| * TN matched WGS | |||
| The datatype is judged by whether the bed file is set (i.e. the `regions` in inputs). | |||
| ### New Releases | |||
| * The TNhaplotyper, named as TNseq in `v0.1.0`, has beed substituted by TNhaplotyper2. | |||
| * The `corealigner` step has been removed. | |||
| * The `vcf2maf` step has been removed. Thus, the final output is the annotated `VCF`. | |||
| ### Getting Started | |||
| We recommend using choppy system and Aliyun OSS service. The command will look like this: | |||
| ``` | |||
| # Activate the choppy environment | |||
| $ open-choppy-env | |||
| # Install the APP | |||
| $ choppy install YaqingLiu/variant-calling-latest [-f] | |||
| # List the parameters | |||
| $ choppy samples YaqingLiu/variant-calling-latest [--no-default] | |||
| # Submit you task with the `samples.csv file` and `project name` | |||
| $ choppy batch YaqingLiu/variant-calling-latest samples.csv -p Project [-l project:Label] | |||
| # Query the status of all tasks in the project | |||
| $ choppy query -L Label | grep "status" | |||
| ``` | |||
| **Please note:** The `defaults` can be forcibly replaced by the settings in `samples.csv`. Therefore, there is no need to contact me over this issue. | |||
| The parameters that must need contains: sample_id,normal_fastq_1,normal_fastq_2,tumor_fastq_1,tumor_fastq_2 | |||
| **Please carefully check** | |||
| * the reference genome | |||
| * bed file | |||
| * the caller you want to use | |||
| * whether PoN needs to be set | |||
BIN
assets/%5CUsers%5Cthink%5CAppData%5CRoaming%5CTypora%5Ctypora-user-images%5C1547626023006.png
Ver arquivo
BIN
assets/1549784133993.png
Ver arquivo
BIN
assets/wgs-germline.png
Ver arquivo
+ 28
- 0
defaults
Ver arquivo
| @@ -0,0 +1,28 @@ | |||
| { | |||
| "fasta": "GRCh38.d1.vd1.fa", | |||
| "ref_dir": "oss://pgx-reference-data/GRCh38.d1.vd1/", | |||
| "dbsnp": "dbsnp_146.hg38.vcf", | |||
| "dbsnp_dir": "oss://pgx-reference-data/GRCh38.d1.vd1/", | |||
| "SENTIEON_INSTALL_DIR": "/opt/sentieon-genomics", | |||
| "SENTIEON_LICENSE": "192.168.0.55:8990", | |||
| "dbmills_dir": "oss://pgx-reference-data/GRCh38.d1.vd1/", | |||
| "db_mills": "Mills_and_1000G_gold_standard.indels.hg38.vcf", | |||
| "germline_resource": "oss://pgx-reference-data/gnomAD/af-only-gnomad.v3.1.1.vcf.gz", | |||
| "sentieon_docker": "registry.cn-shanghai.aliyuncs.com/pgx-docker-registry/sentieon-genomics:v2020.10.07", | |||
| "varscan_docker": "registry.cn-shanghai.aliyuncs.com/pgx-docker-registry/varscan2:v2.4.3", | |||
| "annovar_docker": "registry.cn-shanghai.aliyuncs.com/pgx-docker-registry/annovar:v2018.04", | |||
| "maftools_docker": "registry.cn-shanghai.aliyuncs.com/pgx-docker-registry/r-base:4.0.2", | |||
| "bcftools_docker": "registry.cn-shanghai.aliyuncs.com/pgx-docker-registry/bcftools:v1.9", | |||
| "database": "oss://pgx-reference-data/annovar_hg38/", | |||
| "regions": "oss://pgx-reference-data/bed/cbcga/S07604514_Covered.bed", | |||
| "tnseq_pon": "", | |||
| "tnscope_pon": "", | |||
| "cosmic_vcf": "CosmicCodingMuts.hg38.v91.vcf", | |||
| "cosmic_dir": "oss://pgx-reference-data/reference/cosmic/", | |||
| "disk_size": "200", | |||
| "cluster_config": "OnDemand bcs.a2.3xlarge img-ubuntu-vpc", | |||
| "germline": false, | |||
| "tnseq": true, | |||
| "tnscope": true, | |||
| "varscan": true | |||
| } | |||
+ 32
- 15
inputs
Ver arquivo
| @@ -1,17 +1,34 @@ | |||
| { | |||
| "{{ project_name }}.fasta": "GRCh38.d1.vd1.fa", | |||
| "{{ project_name }}.ref_dir": "oss://pgx-reference-data/GRCh38.d1.vd1/", | |||
| "{{ project_name }}.dbsnp": "dbsnp_146.hg38.vcf", | |||
| "{{ project_name }}.recaled_bam": "{{ recaled_bam }}", | |||
| "{{ project_name }}.SENTIEON_INSTALL_DIR": "/opt/sentieon-genomics", | |||
| "{{ project_name }}.db_mills": "Mills_and_1000G_gold_standard.indels.hg38.vcf", | |||
| "{{ project_name }}.cluster_config": "OnDemand bcs.a2.3xlarge img-ubuntu-vpc", | |||
| "{{ project_name }}.docker": "registry.cn-shanghai.aliyuncs.com/pgx-docker-registry/sentieon-genomics:v2020.10.07", | |||
| "{{ project_name }}.dbsnp_dir": "oss://pgx-reference-data/GRCh38.d1.vd1/", | |||
| "{{ project_name }}.sample": "{{ sample_id }}", | |||
| "{{ project_name }}.disk_size": "500", | |||
| "{{ project_name }}.recaled_bam_index": "{{ recaled_bam_index }}", | |||
| "{{ project_name }}.regions": "oss://pgx-reference-data/bed/cbcga/S07604514_Covered.bed", | |||
| "{{ project_name }}.interval_padding": "0" | |||
| "{{ project_name }}.sample_id": "{{ sample_id }}", | |||
| "{{ project_name }}.tumor_bam": "{{ tumor_bam }}", | |||
| "{{ project_name }}.tumor_bai": "{{ tumor_bai }}", | |||
| "{{ project_name }}.tumor_table": "{{ tumor_table }}", | |||
| "{{ project_name }}.normal_bam": "{{ normal_bam }}", | |||
| "{{ project_name }}.normal_bai": "{{ normal_bai }}", | |||
| "{{ project_name }}.normal_table": "{{ normal_table }}", | |||
| "{{ project_name }}.fasta": "{{ fasta }}", | |||
| "{{ project_name }}.ref_dir": "{{ ref_dir }}", | |||
| "{{ project_name }}.dbsnp": "{{ dbsnp }}", | |||
| "{{ project_name }}.dbsnp_dir": "{{ dbsnp_dir }}", | |||
| "{{ project_name }}.SENTIEON_INSTALL_DIR": "{{ SENTIEON_INSTALL_DIR }}", | |||
| "{{ project_name }}.SENTIEON_LICENSE": "{{ SENTIEON_LICENSE }}", | |||
| "{{ project_name }}.dbmills_dir": "{{ dbmills_dir }}", | |||
| "{{ project_name }}.db_mills": "{{ db_mills }}", | |||
| "{{ project_name }}.germline_resource": "{{ germline_resource }}", | |||
| "{{ project_name }}.sentieon_docker": "{{ sentieon_docker }}", | |||
| "{{ project_name }}.varscan_docker": "{{ varscan_docker }}", | |||
| "{{ project_name }}.annovar_docker": "{{ annovar_docker }}", | |||
| "{{ project_name }}.maftools_docker": "{{ maftools_docker }}", | |||
| "{{ project_name }}.database": "{{ database }}", | |||
| "{{ project_name }}.regions": "{{ regions }}", | |||
| "{{ project_name }}.tnseq_pon": "{{ tnseq_pon }}", | |||
| "{{ project_name }}.tnscope_pon": "{{ tnscope_pon }}", | |||
| "{{ project_name }}.cosmic_vcf": "{{ cosmic_vcf }}", | |||
| "{{ project_name }}.cosmic_dir": "{{ cosmic_dir }}", | |||
| "{{ project_name }}.disk_size": "{{ disk_size }}", | |||
| "{{ project_name }}.cluster_config": "{{ cluster_config }}", | |||
| "{{ project_name }}.germline": {{ germline | tojson }}, | |||
| "{{ project_name }}.tnseq": {{ tnseq | tojson }}, | |||
| "{{ project_name }}.tnscope": {{ tnscope | tojson }}, | |||
| "{{ project_name }}.varscan": {{ varscan | tojson }} | |||
| } | |||
+ 0
- 48
tasks/BQSR.wdl
Ver arquivo
| @@ -1,48 +0,0 @@ | |||
| task BQSR { | |||
| File ref_dir | |||
| File dbsnp_dir | |||
| File dbmills_dir | |||
| String sample | |||
| String SENTIEON_INSTALL_DIR | |||
| String fasta | |||
| String dbsnp | |||
| String db_mills | |||
| File realigned_bam | |||
| File realigned_bam_index | |||
| String docker | |||
| String cluster_config | |||
| String disk_size | |||
| command <<< | |||
| set -o pipefail | |||
| set -e | |||
| export SENTIEON_LICENSE=192.168.0.55:8990 | |||
| nt=$(nproc) | |||
| ${SENTIEON_INSTALL_DIR}/bin/sentieon driver -r ${ref_dir}/${fasta} -t $nt -i ${realigned_bam} --algo QualCal -k ${dbsnp_dir}/${dbsnp} -k ${dbmills_dir}/${db_mills} ${sample}_recal_data.table | |||
| ${SENTIEON_INSTALL_DIR}/bin/sentieon driver -r ${ref_dir}/${fasta} -t $nt -i ${realigned_bam} -q ${sample}_recal_data.table --algo QualCal -k ${dbsnp_dir}/${dbsnp} -k ${dbmills_dir}/${db_mills} ${sample}_recal_data.table.post --algo ReadWriter ${sample}.sorted.deduped.realigned.recaled.bam | |||
| ${SENTIEON_INSTALL_DIR}/bin/sentieon driver -t $nt --algo QualCal --plot --before ${sample}_recal_data.table --after ${sample}_recal_data.table.post ${sample}_recal_data.csv | |||
| ${SENTIEON_INSTALL_DIR}/bin/sentieon plot QualCal -o ${sample}_bqsrreport.pdf ${sample}_recal_data.csv | |||
| >>> | |||
| runtime { | |||
| docker:docker | |||
| cluster: cluster_config | |||
| systemDisk: "cloud_ssd 40" | |||
| dataDisk: "cloud_ssd " + disk_size + " /cromwell_root/" | |||
| } | |||
| output { | |||
| File recal_table = "${sample}_recal_data.table" | |||
| File recal_post = "${sample}_recal_data.table.post" | |||
| File recaled_bam = "${sample}.sorted.deduped.realigned.recaled.bam" | |||
| File recaled_bam_index = "${sample}.sorted.deduped.realigned.recaled.bam.bai" | |||
| File recal_csv = "${sample}_recal_data.csv" | |||
| File bqsrreport_pdf = "${sample}_bqsrreport.pdf" | |||
| } | |||
| } | |||
+ 0
- 40
tasks/Dedup.wdl
Ver arquivo
| @@ -1,40 +0,0 @@ | |||
| task Dedup { | |||
| String SENTIEON_INSTALL_DIR | |||
| String sample | |||
| File sorted_bam | |||
| File sorted_bam_index | |||
| String docker | |||
| String cluster_config | |||
| String disk_size | |||
| command <<< | |||
| set -o pipefail | |||
| set -e | |||
| export SENTIEON_LICENSE=192.168.0.55:8990 | |||
| nt=$(nproc) | |||
| ${SENTIEON_INSTALL_DIR}/bin/sentieon driver -t $nt -i ${sorted_bam} --algo LocusCollector --fun score_info ${sample}_score.txt | |||
| ${SENTIEON_INSTALL_DIR}/bin/sentieon driver -t $nt -i ${sorted_bam} --algo Dedup --rmdup --score_info ${sample}_score.txt --metrics ${sample}_dedup_metrics.txt ${sample}.sorted.deduped.bam | |||
| >>> | |||
| runtime { | |||
| docker:docker | |||
| cluster: cluster_config | |||
| systemDisk: "cloud_ssd 40" | |||
| dataDisk: "cloud_ssd " + disk_size + " /cromwell_root/" | |||
| } | |||
| output { | |||
| File score = "${sample}_score.txt" | |||
| File dedup_metrics = "${sample}_dedup_metrics.txt" | |||
| File Dedup_bam = "${sample}.sorted.deduped.bam" | |||
| File Dedup_bam_index = "${sample}.sorted.deduped.bam.bai" | |||
| } | |||
| } | |||
+ 0
- 49
tasks/Haplotyper.wdl
Ver arquivo
| @@ -1,49 +0,0 @@ | |||
| task Haplotyper { | |||
| File ref_dir | |||
| File dbsnp_dir | |||
| String SENTIEON_INSTALL_DIR | |||
| String fasta | |||
| File recaled_bam | |||
| File recaled_bam_index | |||
| String dbsnp | |||
| String sample | |||
| String docker | |||
| String cluster_config | |||
| String disk_size | |||
| File? regions | |||
| Int? interval_padding | |||
| command <<< | |||
| set -o pipefail | |||
| set -e | |||
| export SENTIEON_LICENSE=192.168.0.55:8990 | |||
| nt=$(nproc) | |||
| if [ ${regions} ]; then | |||
| INTERVAL="--interval ${regions} --interval_padding ${interval_padding}" | |||
| else | |||
| INTERVAL="" | |||
| fi | |||
| ${SENTIEON_INSTALL_DIR}/bin/sentieon driver -r ${ref_dir}/${fasta} -t $nt \ | |||
| -i ${recaled_bam} $INTERVAL \ | |||
| --algo Haplotyper -d ${dbsnp_dir}/${dbsnp} ${sample}_hc.vcf | |||
| >>> | |||
| runtime { | |||
| docker:docker | |||
| cluster: cluster_config | |||
| systemDisk: "cloud_ssd 40" | |||
| dataDisk: "cloud_ssd " + disk_size + " /cromwell_root/" | |||
| } | |||
| output { | |||
| File vcf = "${sample}_hc.vcf" | |||
| File vcf_idx = "${sample}_hc.vcf.idx" | |||
| } | |||
| } | |||
+ 0
- 56
tasks/Metrics.wdl
Ver arquivo
| @@ -1,56 +0,0 @@ | |||
| task Metrics { | |||
| File ref_dir | |||
| String SENTIEON_INSTALL_DIR | |||
| String sample | |||
| String docker | |||
| String cluster_config | |||
| String fasta | |||
| File sorted_bam | |||
| File sorted_bam_index | |||
| String disk_size | |||
| command <<< | |||
| set -o pipefail | |||
| set -e | |||
| export SENTIEON_LICENSE=192.168.0.55:8990 | |||
| nt=$(nproc) | |||
| ${SENTIEON_INSTALL_DIR}/bin/sentieon driver -r ${ref_dir}/${fasta} -t $nt -i ${sorted_bam} --algo MeanQualityByCycle ${sample}_mq_metrics.txt --algo QualDistribution ${sample}_qd_metrics.txt --algo GCBias --summary ${sample}_gc_summary.txt ${sample}_gc_metrics.txt --algo AlignmentStat ${sample}_aln_metrics.txt --algo InsertSizeMetricAlgo ${sample}_is_metrics.txt --algo CoverageMetrics --omit_base_output ${sample}_coverage_metrics | |||
| ${SENTIEON_INSTALL_DIR}/bin/sentieon plot metrics -o ${sample}_metrics_report.pdf gc=${sample}_gc_metrics.txt qd=${sample}_qd_metrics.txt mq=${sample}_mq_metrics.txt isize=${sample}_is_metrics.txt | |||
| >>> | |||
| runtime { | |||
| docker:docker | |||
| cluster: cluster_config | |||
| systemDisk: "cloud_ssd 40" | |||
| dataDisk: "cloud_ssd " + disk_size + " /cromwell_root/" | |||
| } | |||
| output { | |||
| File qd_metrics = "${sample}_qd_metrics.txt" | |||
| File qd_metrics_pdf = "${sample}_qd_metrics.pdf" | |||
| File mq_metrics = "${sample}_mq_metrics.txt" | |||
| File mq_metrics_pdf = "${sample}_mq_metrics.pdf" | |||
| File is_metrics = "${sample}_is_metrics.txt" | |||
| File is_metrics_pdf = "${sample}_is_metrics.pdf" | |||
| File gc_summary = "${sample}_gc_summary.txt" | |||
| File gc_metrics = "${sample}_gc_metrics.txt" | |||
| File gc_metrics_pdf = "${sample}_gc_metrics.pdf" | |||
| File aln_metrics = "${sample}_aln_metrics.txt" | |||
| File coverage_metrics_sample_summary = "${sample}_coverage_metrics.sample_summary" | |||
| File coverage_metrics_sample_statistics = "${sample}_coverage_metrics.sample_statistics" | |||
| File coverage_metrics_sample_interval_statistics = "${sample}_coverage_metrics.sample_interval_statistics" | |||
| File coverage_metrics_sample_cumulative_coverage_proportions = "${sample}_coverage_metrics.sample_cumulative_coverage_proportions" | |||
| File coverage_metrics_sample_cumulative_coverage_counts = "${sample}_coverage_metrics.sample_cumulative_coverage_counts" | |||
| } | |||
| } | |||
+ 0
- 41
tasks/Realigner.wdl
Ver arquivo
| @@ -1,41 +0,0 @@ | |||
| task Realigner { | |||
| File ref_dir | |||
| File dbmills_dir | |||
| String SENTIEON_INSTALL_DIR | |||
| String sample | |||
| String fasta | |||
| File Dedup_bam | |||
| File Dedup_bam_index | |||
| String db_mills | |||
| String docker | |||
| String cluster_config | |||
| String disk_size | |||
| command <<< | |||
| set -o pipefail | |||
| set -e | |||
| export SENTIEON_LICENSE=192.168.0.55:8990 | |||
| nt=$(nproc) | |||
| ${SENTIEON_INSTALL_DIR}/bin/sentieon driver -r ${ref_dir}/${fasta} -t $nt -i ${Dedup_bam} --algo Realigner -k ${dbmills_dir}/${db_mills} ${sample}.sorted.deduped.realigned.bam | |||
| >>> | |||
| runtime { | |||
| docker:docker | |||
| cluster: cluster_config | |||
| systemDisk: "cloud_ssd 40" | |||
| dataDisk: "cloud_ssd " + disk_size + " /cromwell_root/" | |||
| } | |||
| output { | |||
| File realigner_bam = "${sample}.sorted.deduped.realigned.bam" | |||
| File realigner_bam_index = "${sample}.sorted.deduped.realigned.bam.bai" | |||
| } | |||
| } | |||
+ 63
- 36
tasks/TNscope.wdl
Ver arquivo
| @@ -1,42 +1,69 @@ | |||
| task TNscope { | |||
| String sample | |||
| String SENTIEON_INSTALL_DIR | |||
| String SENTIEON_LICENSE | |||
| File tumor_recaled_bam | |||
| File tumor_recaled_bam_index | |||
| File tumor_recal_table | |||
| File normal_recaled_bam | |||
| File normal_recaled_bam_index | |||
| File normal_recal_table | |||
| String tumor_name | |||
| String normal_name | |||
| File ref_dir | |||
| File dbsnp_dir | |||
| File ref_dir | |||
| String fasta | |||
| File dbsnp_dir | |||
| String dbsnp | |||
| File? regions | |||
| Int? interval_padding | |||
| File? pon_vcf | |||
| String docker | |||
| String cluster_config | |||
| String disk_size | |||
| command <<< | |||
| set -o pipefail | |||
| set -e | |||
| export SENTIEON_LICENSE=${SENTIEON_LICENSE} | |||
| nt=$(nproc) | |||
| if [ ${regions} ]; then | |||
| INTERVAL="--interval ${regions} --interval_padding ${interval_padding}" | |||
| else | |||
| INTERVAL="" | |||
| fi | |||
| if [ ${pon_vcf} ]; then | |||
| PON="--pon ${pon_vcf}" | |||
| ${SENTIEON_INSTALL_DIR}/bin/sentieon util vcfindex ${pon_vcf} | |||
| else | |||
| PON="" | |||
| fi | |||
| ${SENTIEON_INSTALL_DIR}/bin/sentieon driver -t $nt \ | |||
| -r ${ref_dir}/${fasta} $INTERVAL \ | |||
| -i ${tumor_recaled_bam} -q ${tumor_recal_table} \ | |||
| -i ${normal_recaled_bam} -q ${normal_recal_table} \ | |||
| --algo TNscope \ | |||
| --tumor_sample ${tumor_name} --normal_sample ${normal_name} \ | |||
| --dbsnp ${dbsnp_dir}/${dbsnp} \ | |||
| $PON \ | |||
| ${sample}.TNscope.TN.vcf | |||
| >>> | |||
| runtime { | |||
| docker: docker | |||
| cluster: cluster_config | |||
| systemDisk: "cloud_ssd 40" | |||
| dataDisk: "cloud_ssd " + disk_size + " /cromwell_root/" | |||
| } | |||
| String SENTIEON_INSTALL_DIR | |||
| String tumor_name | |||
| String normal_name | |||
| String docker | |||
| String cluster_config | |||
| String fasta | |||
| File corealigner_bam | |||
| File corealigner_bam_index | |||
| String dbsnp | |||
| String disk_size | |||
| command <<< | |||
| set -o pipefail | |||
| set -e | |||
| export SENTIEON_LICENSE=192.168.0.55:8990 | |||
| nt=$(nproc) | |||
| ${SENTIEON_INSTALL_DIR}/bin/sentieon driver -r ${ref_dir}/${fasta} -t $nt -i ${corealigner_bam} --algo TNscope --tumor_sample ${tumor_name} --normal_sample ${normal_name} --dbsnp ${dbsnp_dir}/${dbsnp} ${sample}.TNscope.TN.vcf | |||
| >>> | |||
| runtime { | |||
| docker:docker | |||
| cluster: cluster_config | |||
| systemDisk: "cloud_ssd 40" | |||
| dataDisk: "cloud_ssd " + disk_size + " /cromwell_root/" | |||
| } | |||
| output { | |||
| File TNscope_vcf= "${sample}.TNscope.TN.vcf" | |||
| File TNscope_vcf_index = "${sample}.TNscope.TN.vcf.idx" | |||
| } | |||
| output { | |||
| File TNscope_vcf= "${sample}.TNscope.TN.vcf" | |||
| File TNscope_vcf_index = "${sample}.TNscope.TN.vcf.idx" | |||
| } | |||
| } | |||
+ 86
- 40
tasks/TNseq.wdl
Ver arquivo
| @@ -1,43 +1,89 @@ | |||
| task TNseq { | |||
| File ref_dir | |||
| File dbsnp_dir | |||
| String SENTIEON_INSTALL_DIR | |||
| String tumor_name | |||
| String normal_name | |||
| String docker | |||
| String cluster_config | |||
| String fasta | |||
| File corealigner_bam | |||
| File corealigner_bam_index | |||
| String dbsnp | |||
| String disk_size | |||
| command <<< | |||
| set -o pipefail | |||
| set -e | |||
| export SENTIEON_LICENSE=192.168.0.55:8990 | |||
| nt=$(nproc) | |||
| ${SENTIEON_INSTALL_DIR}/bin/sentieon driver -r ${ref_dir}/${fasta} -t $nt -i ${corealigner_bam} --algo TNhaplotyper --tumor_sample ${tumor_name} --normal_sample ${normal_name} --dbsnp ${dbsnp_dir}/${dbsnp} ${sample}.TNseq.TN.vcf | |||
| >>> | |||
| runtime { | |||
| docker:docker | |||
| cluster: cluster_config | |||
| systemDisk: "cloud_ssd 40" | |||
| dataDisk: "cloud_ssd " + disk_size + " /cromwell_root/" | |||
| } | |||
| output { | |||
| File TNseq_vcf= "${sample}.TNseq.TN.vcf" | |||
| File TNseq_vcf_index = "${sample}.TNseq.TN.vcf.idx" | |||
| } | |||
| } | |||
| String sample | |||
| String SENTIEON_INSTALL_DIR | |||
| String SENTIEON_LICENSE | |||
| File tumor_recaled_bam | |||
| File tumor_recaled_bam_index | |||
| File tumor_recal_table | |||
| File normal_recaled_bam | |||
| File normal_recaled_bam_index | |||
| File normal_recal_table | |||
| String tumor_name | |||
| String normal_name | |||
| File ref_dir | |||
| String fasta | |||
| File germline_resource | |||
| File? regions | |||
| Int? interval_padding | |||
| File? pon_vcf | |||
| String docker | |||
| String cluster_config | |||
| String disk_size | |||
| command <<< | |||
| set -o pipefail | |||
| set -e | |||
| export SENTIEON_LICENSE=${SENTIEON_LICENSE} | |||
| nt=$(nproc) | |||
| if [ ${regions} ]; then | |||
| INTERVAL="--interval ${regions} --interval_padding ${interval_padding}" | |||
| else | |||
| INTERVAL="" | |||
| fi | |||
| if [ ${pon_vcf} ]; then | |||
| PON="--pon ${pon_vcf}" | |||
| ${SENTIEON_INSTALL_DIR}/bin/sentieon util vcfindex ${pon_vcf} | |||
| else | |||
| PON="" | |||
| fi | |||
| ${SENTIEON_INSTALL_DIR}/bin/sentieon driver -t $nt \ | |||
| -r ${ref_dir}/${fasta} $INTERVAL \ | |||
| -i ${tumor_recaled_bam} -q ${tumor_recal_table} \ | |||
| -i ${normal_recaled_bam} -q ${normal_recal_table} \ | |||
| --algo TNhaplotyper2 \ | |||
| --tumor_sample ${tumor_name} --normal_sample ${normal_name} \ | |||
| --germline_vcf ${germline_resource} \ | |||
| $PON \ | |||
| ${sample}.TNseq.TN.tmp.vcf \ | |||
| --algo OrientationBias --tumor_sample ${tumor_name} \ | |||
| ${sample}.orientation \ | |||
| --algo ContaminationModel \ | |||
| --tumor_sample ${tumor_name} --normal_sample ${normal_name} \ | |||
| --vcf ${germline_resource} \ | |||
| --tumor_segments ${sample}.contamination.segments \ | |||
| ${sample}.contamination | |||
| sentieon driver -r REFERENCE \ | |||
| --algo TNfilter \ | |||
| --tumor_sample ${tumor_name} --normal_sample ${normal_name} \ | |||
| -v ${sample}.TNseq.TN.tmp.vcf \ | |||
| --contamination ${sample}.contamination \ | |||
| --tumor_segments ${sample}.contamination.segments \ | |||
| --orientation_priors ${sample}.orientation \ | |||
| ${sample}.TNseq.TN.vcf | |||
| >>> | |||
| runtime { | |||
| docker: docker | |||
| cluster: cluster_config | |||
| systemDisk: "cloud_ssd 40" | |||
| dataDisk: "cloud_ssd " + disk_size + " /cromwell_root/" | |||
| } | |||
| output { | |||
| File TNseq_vcf = "${sample}.TNseq.TN.vcf" | |||
| File TNseq_vcf_index = "${sample}.TNseq.TN.vcf.idx" | |||
| File contamination = "${sample}.contamination" | |||
| File contamination_segments = "${sample}.contamination.segments" | |||
| File orientation = "${sample}.orientation" | |||
| } | |||
| } | |||
+ 0
- 44
tasks/corealigner.wdl
Ver arquivo
| @@ -1,44 +0,0 @@ | |||
| task corealigner { | |||
| File ref_dir | |||
| File dbsnp_dir | |||
| File dbmills_dir | |||
| String sample | |||
| String SENTIEON_INSTALL_DIR | |||
| String docker | |||
| String cluster_config | |||
| String fasta | |||
| String dbsnp | |||
| String db_mills | |||
| File tumor_recaled_bam | |||
| File tumor_recaled_bam_index | |||
| File normal_recaled_bam | |||
| File normal_recaled_bam_index | |||
| String disk_size | |||
| command <<< | |||
| set -o pipefail | |||
| set -e | |||
| export SENTIEON_LICENSE=192.168.0.55:8990 | |||
| nt=$(nproc) | |||
| ${SENTIEON_INSTALL_DIR}/bin/sentieon driver -r ${ref_dir}/${fasta} -t $nt -i ${tumor_recaled_bam} -i ${normal_recaled_bam} --algo Realigner -k ${db_mills} -k ${dbsnp} ${sample}_corealigned.bam | |||
| >>> | |||
| runtime { | |||
| docker:docker | |||
| cluster: cluster_config | |||
| systemDisk: "cloud_ssd 40" | |||
| dataDisk: "cloud_ssd " + disk_size + " /cromwell_root/" | |||
| } | |||
| output { | |||
| File corealigner_bam = "${sample}_corealigned.bam" | |||
| File corealigner_bam_index = "${sample}_corealigned.bam.bai" | |||
| } | |||
| } | |||
+ 0
- 37
tasks/deduped_Metrics.wdl
Ver arquivo
| @@ -1,37 +0,0 @@ | |||
| task deduped_Metrics { | |||
| File ref_dir | |||
| String SENTIEON_INSTALL_DIR | |||
| String sample | |||
| String fasta | |||
| File Dedup_bam | |||
| File Dedup_bam_index | |||
| String docker | |||
| String cluster_config | |||
| String disk_size | |||
| command <<< | |||
| set -o pipefail | |||
| set -e | |||
| export SENTIEON_LICENSE=192.168.0.55:8990 | |||
| nt=$(nproc) | |||
| ${SENTIEON_INSTALL_DIR}/bin/sentieon driver -r ${ref_dir}/${fasta} -t $nt -i ${Dedup_bam} --algo CoverageMetrics --omit_base_output ${sample}_deduped_coverage_metrics --algo MeanQualityByCycle ${sample}_deduped_mq_metrics.txt --algo QualDistribution ${sample}_deduped_qd_metrics.txt --algo GCBias --summary ${sample}_deduped_gc_summary.txt ${sample}_deduped_gc_metrics.txt --algo AlignmentStat ${sample}_deduped_aln_metrics.txt --algo InsertSizeMetricAlgo ${sample}_deduped_is_metrics.txt | |||
| >>> | |||
| runtime { | |||
| docker:docker | |||
| cluster: cluster_config | |||
| systemDisk: "cloud_ssd 40" | |||
| dataDisk: "cloud_ssd " + disk_size + " /cromwell_root/" | |||
| } | |||
| output { | |||
| File deduped_coverage_metrics_sample_summary = "${sample}_deduped_coverage_metrics.sample_summary" | |||
| File deduped_coverage_metrics_sample_statistics = "${sample}_deduped_coverage_metrics.sample_statistics" | |||
| File deduped_coverage_metrics_sample_interval_statistics = "${sample}_deduped_coverage_metrics.sample_interval_statistics" | |||
| File deduped_coverage_metrics_sample_cumulative_coverage_proportions = "${sample}_deduped_coverage_metrics.sample_cumulative_coverage_proportions" | |||
| File deduped_coverage_metrics_sample_cumulative_coverage_counts = "${sample}_deduped_coverage_metrics.sample_cumulative_coverage_counts" | |||
| } | |||
| } | |||
+ 0
- 34
tasks/mapping.wdl
Ver arquivo
| @@ -1,34 +0,0 @@ | |||
| task mapping { | |||
| File ref_dir | |||
| String fasta | |||
| File fastq_1 | |||
| File fastq_2 | |||
| String SENTIEON_INSTALL_DIR | |||
| String group | |||
| String sample | |||
| String pl | |||
| String docker | |||
| String cluster_config | |||
| String disk_size | |||
| command <<< | |||
| set -o pipefail | |||
| set -e | |||
| export SENTIEON_LICENSE=192.168.0.55:8990 | |||
| nt=$(nproc) | |||
| ${SENTIEON_INSTALL_DIR}/bin/bwa mem -M -R "@RG\tID:${group}\tSM:${sample}\tPL:${pl}" -t $nt -K 10000000 ${ref_dir}/${fasta} ${fastq_1} ${fastq_2} | ${SENTIEON_INSTALL_DIR}/bin/sentieon util sort -o ${sample}.sorted.bam -t $nt --sam2bam -i - | |||
| >>> | |||
| runtime { | |||
| docker:docker | |||
| cluster: cluster_config | |||
| systemDisk: "cloud_ssd 40" | |||
| dataDisk: "cloud_ssd " + disk_size + " /cromwell_root/" | |||
| } | |||
| output { | |||
| File sorted_bam = "${sample}.sorted.bam" | |||
| File sorted_bam_index = "${sample}.sorted.bam.bai" | |||
| } | |||
| } | |||
+ 92
- 33
workflow.wdl
Ver arquivo
| @@ -1,39 +1,98 @@ | |||
| import "./tasks/Haplotyper.wdl" as Haplotyper | |||
| import "./tasks/TNseq.wdl" as TNseq | |||
| import "./tasks/TNscope.wdl" as TNscope | |||
| workflow {{ project_name }} { | |||
| String sample_id | |||
| File tumor_bam | |||
| File tumor_bai | |||
| File tumor_table | |||
| File normal_bam | |||
| File normal_bai | |||
| File normal_table | |||
| String SENTIEON_INSTALL_DIR | |||
| String SENTIEON_LICENSE | |||
| String sentieon_docker | |||
| String varscan_docker | |||
| String annovar_docker | |||
| String maftools_docker | |||
| File ref_dir | |||
| String fasta | |||
| File dbmills_dir | |||
| String db_mills | |||
| File dbsnp_dir | |||
| String dbsnp | |||
| File germline_resource | |||
| File? regions | |||
| Int? interval_padding | |||
| File database | |||
| String disk_size | |||
| String cluster_config | |||
| File recaled_bam | |||
| File recaled_bam_index | |||
| File? tnseq_pon | |||
| File? tnscope_pon | |||
| File? cosmic_dir | |||
| String? cosmic_vcf | |||
| String SENTIEON_INSTALL_DIR | |||
| String sample | |||
| String docker | |||
| String fasta | |||
| File ref_dir | |||
| File dbsnp_dir | |||
| String dbsnp | |||
| String disk_size | |||
| String cluster_config | |||
| Boolean germline | |||
| Boolean tnseq | |||
| Boolean tnscope | |||
| Boolean varscan | |||
| File? regions | |||
| Int? interval_padding | |||
| if (tnseq) { | |||
| call TNseq.TNseq as TNseq { | |||
| input: | |||
| SENTIEON_INSTALL_DIR=SENTIEON_INSTALL_DIR, | |||
| SENTIEON_LICENSE=SENTIEON_LICENSE, | |||
| sample=sample_id, | |||
| normal_recaled_bam=normal_bam, | |||
| normal_recaled_bam_index=normal_bai, | |||
| normal_recal_table=normal_table, | |||
| tumor_recaled_bam=tumor_bam, | |||
| tumor_recaled_bam_index=tumor_bai, | |||
| tumor_recal_table=tumor_table, | |||
| normal_name=sample_id + "_normal", | |||
| tumor_name=sample_id + "_tumor", | |||
| fasta=fasta, | |||
| ref_dir=ref_dir, | |||
| regions=regions, | |||
| interval_padding=interval_padding, | |||
| germline_resource=germline_resource, | |||
| pon_vcf=tnseq_pon, | |||
| docker=sentieon_docker, | |||
| cluster_config=cluster_config, | |||
| disk_size=disk_size | |||
| } | |||
| } | |||
| call Haplotyper.Haplotyper as Haplotyper { | |||
| input: | |||
| SENTIEON_INSTALL_DIR=SENTIEON_INSTALL_DIR, | |||
| fasta=fasta, | |||
| ref_dir=ref_dir, | |||
| regions=regions, | |||
| interval_padding=interval_padding, | |||
| recaled_bam=recaled_bam, | |||
| recaled_bam_index=recaled_bam_index, | |||
| dbsnp=dbsnp, | |||
| dbsnp_dir=dbsnp_dir, | |||
| sample=sample, | |||
| docker=docker, | |||
| disk_size=disk_size, | |||
| cluster_config=cluster_config | |||
| } | |||
| } | |||
| if (tnscope) { | |||
| call TNscope.TNscope as TNscope { | |||
| input: | |||
| SENTIEON_INSTALL_DIR=SENTIEON_INSTALL_DIR, | |||
| SENTIEON_LICENSE=SENTIEON_LICENSE, | |||
| sample=sample_id, | |||
| normal_recaled_bam=normal_bam, | |||
| normal_recaled_bam_index=normal_bai, | |||
| normal_recal_table=normal_table, | |||
| tumor_recaled_bam=tumor_bam, | |||
| tumor_recaled_bam_index=tumor_bai, | |||
| tumor_recal_table=tumor_table, | |||
| normal_name=sample_id + "_normal", | |||
| tumor_name=sample_id + "_tumor", | |||
| fasta=fasta, | |||
| ref_dir=ref_dir, | |||
| regions=regions, | |||
| interval_padding=interval_padding, | |||
| dbsnp=dbsnp, | |||
| dbsnp_dir=dbsnp_dir, | |||
| pon_vcf=tnscope_pon, | |||
| docker=sentieon_docker, | |||
| cluster_config=cluster_config, | |||
| disk_size=disk_size | |||
| } | |||
| } | |||
| } | |||
Carregando…