işleme
28cf29e79e
24 değiştirilmiş dosya ile 989 ekleme ve 0 silme
+ 42
- 0
README.md
Dosyayı Görüntüle
| @@ -0,0 +1,42 @@ | |||
| #高置信位点整合Part2——从VCF和BAM文件中提取突变位点的信息 | |||
| > Author: Run Luyao | |||
| > | |||
| > E-mail:18110700050@fudan.edu.cn | |||
| > | |||
| > Git: http://choppy.3steps.cn/renluyao/HCC_Extract_Info.git | |||
| > | |||
| > Last Updates: 11/9/2019 | |||
| ## 安装指南 | |||
| ```bash | |||
| # 激活choppy环境 | |||
| source activate choppy | |||
| # 安装app | |||
| choppy install renluyao/HCC_Extract_Info | |||
| ``` | |||
| ## App概述 | |||
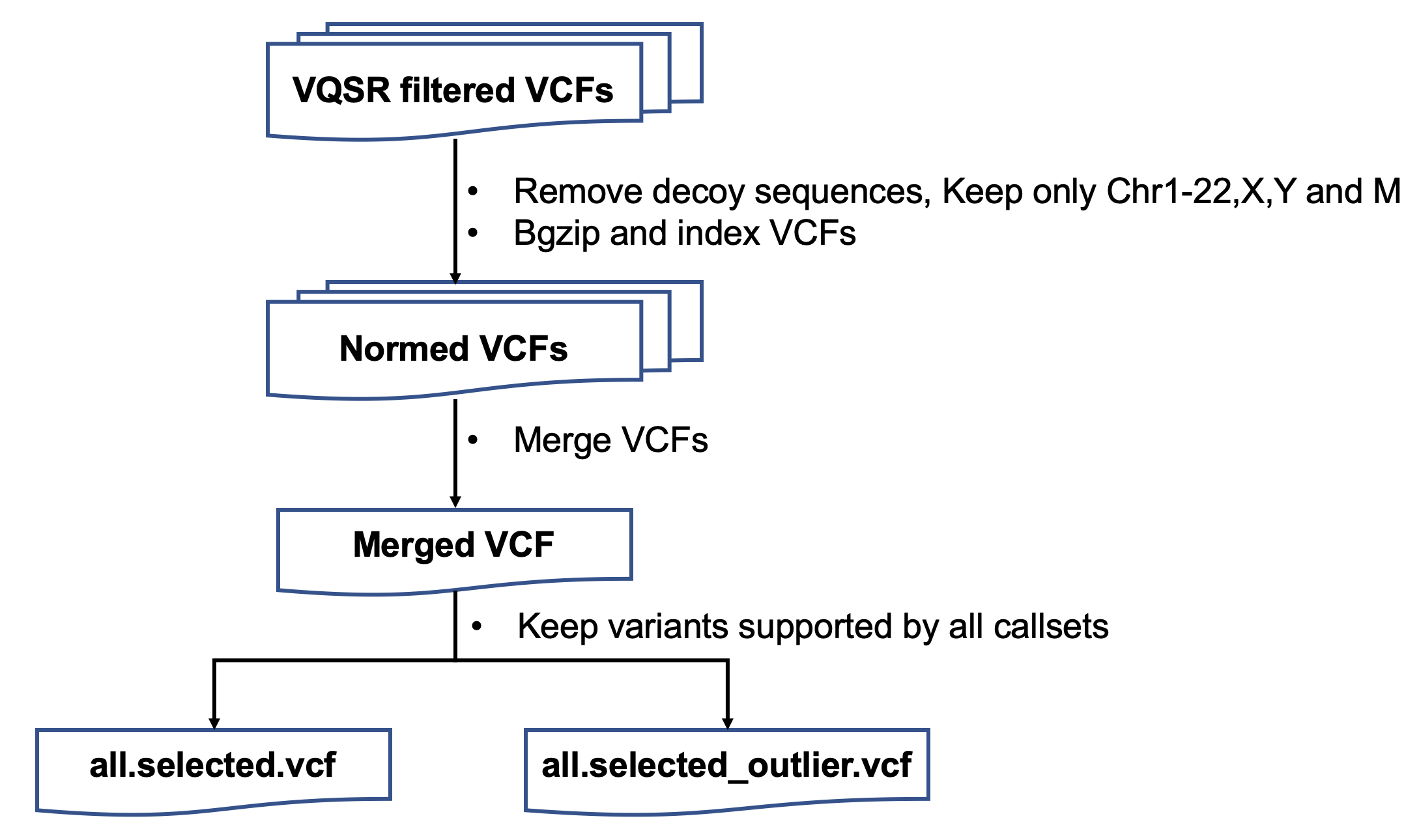
| 当我们拿到测序结果的时候,没有办法判断哪些是真的突变位点,哪些是假阳性 | |||
| ## 流程与参数 | |||
| 两个问题: | |||
| 1. MNP | |||
| 2. merge之后的pos相同,但是ref和alt不同 | |||
| 是不是gatk的问题,尝试用bcftools norm | |||
| ## App输入变量与输入文件 | |||
| ## App输出文件 | |||
| ## 结果展示与解读 | |||
| ## CHANGELOG | |||
| ## FAQ | |||
BIN
codescripts/.DS_Store
Dosyayı Görüntüle
+ 68
- 0
codescripts/bed_for_bamReadcount.py
Dosyayı Görüntüle
| @@ -0,0 +1,68 @@ | |||
| import sys,getopt | |||
| import os | |||
| import re | |||
| import fileinput | |||
| def usage(): | |||
| print( | |||
| """ | |||
| Usage: python bed_for_bamReadcount.py -i input_vcf_file -o prefix | |||
| This script selects SNPs and Indels supported by all callsets. | |||
| Please notice that bam-readcount only takes in 1-based coordinates. | |||
| Input: | |||
| -i a vcf file | |||
| Output: | |||
| -o a indel bed file for bam-readcount | |||
| """) | |||
| # select supported small variants | |||
| def process(oneLine): | |||
| m = re.match('^\#',oneLine) | |||
| if m is not None: | |||
| pass | |||
| else: | |||
| line = oneLine.rstrip() | |||
| strings = line.strip().split('\t') | |||
| # convert the position to bed file for bam-readcount | |||
| # deletion | |||
| if len(strings[3]) > 1 and len(strings[4]) == 1: | |||
| pos = int(strings[1]) + 1 | |||
| outline = strings[0] + '\t' + str(pos) + '\t' + str(pos) + '\t' + strings[3] + '\t' + strings[4]+'\n' | |||
| outINDEL.write(outline) | |||
| # insertion | |||
| elif len(strings[3]) == 1 and len(strings[4]) > 1 and (',' not in strings[4]): | |||
| outline = strings[0] + '\t' + strings[1] + '\t' + strings[1] + '\t' + strings[3] + '\t' + strings[4] + '\n' | |||
| outINDEL.write(outline) | |||
| else: | |||
| outMNP.write(oneLine) | |||
| opts,args = getopt.getopt(sys.argv[1:],"hi:o:") | |||
| for op,value in opts: | |||
| if op == "-i": | |||
| inputFile=value | |||
| elif op == "-o": | |||
| prefix=value | |||
| elif op == "-h": | |||
| usage() | |||
| sys.exit() | |||
| if len(sys.argv[1:]) < 3: | |||
| usage() | |||
| sys.exit() | |||
| INDELname = prefix + '.bed' | |||
| MNPname = prefix + '_MNP.txt' | |||
| outINDEL = open(INDELname,'w') | |||
| outMNP = open(MNPname,'w') | |||
| for line in fileinput.input(inputFile): | |||
| process(line) | |||
| outINDEL.close() | |||
| outMNP.close() | |||
+ 96
- 0
codescripts/extract_vcf_information.py
Dosyayı Görüntüle
| @@ -0,0 +1,96 @@ | |||
| import sys,getopt | |||
| import os | |||
| import re | |||
| import fileinput | |||
| import pandas as pd | |||
| def usage(): | |||
| print( | |||
| """ | |||
| Usage: python extract_vcf_information.py -i input_merged_vcf_file -o parsed_file | |||
| This script will extract SNVs and Indels information from the vcf files and output a tab-delimited files. | |||
| Input: | |||
| -i the selected vcf file | |||
| Output: | |||
| -o tab-delimited parsed file | |||
| """) | |||
| # select supported small variants | |||
| def process(oneLine): | |||
| line = oneLine.rstrip() | |||
| strings = line.strip().split('\t') | |||
| infoParsed = parse_INFO(strings[7]) | |||
| formatKeys = strings[8].split(':') | |||
| formatValues = strings[9].split(':') | |||
| for i in range(0,len(formatKeys) -1) : | |||
| if formatKeys[i] == 'AD': | |||
| ra = formatValues[i].split(',') | |||
| infoParsed['RefDP'] = ra[0] | |||
| infoParsed['AltDP'] = ra[1] | |||
| if (int(ra[1]) + int(ra[0])) != 0: | |||
| infoParsed['af'] = float(int(ra[1])/(int(ra[1]) + int(ra[0]))) | |||
| else: | |||
| pass | |||
| else: | |||
| infoParsed[formatKeys[i]] = formatValues[i] | |||
| infoParsed['chromo'] = strings[0] | |||
| infoParsed['pos'] = strings[1] | |||
| infoParsed['id'] = strings[2] | |||
| infoParsed['ref'] = strings[3] | |||
| infoParsed['alt'] = strings[4] | |||
| infoParsed['qual'] = strings[5] | |||
| return infoParsed | |||
| def parse_INFO(info): | |||
| strings = info.strip().split(';') | |||
| keys = [] | |||
| values = [] | |||
| for i in strings: | |||
| kv = i.split('=') | |||
| if kv[0] == 'DB': | |||
| keys.append('DB') | |||
| values.append('1') | |||
| elif kv[0] == 'AF': | |||
| pass | |||
| elif kv[0] == 'POSITIVE_TRAIN_SITE': | |||
| pass | |||
| elif kv[0] == 'NEGATIVE_TRAIN_SITE': | |||
| pass | |||
| else: | |||
| keys.append(kv[0]) | |||
| values.append(kv[1]) | |||
| infoDict = dict(zip(keys, values)) | |||
| return infoDict | |||
| opts,args = getopt.getopt(sys.argv[1:],"hi:o:") | |||
| for op,value in opts: | |||
| if op == "-i": | |||
| inputFile=value | |||
| elif op == "-o": | |||
| outputFile=value | |||
| elif op == "-h": | |||
| usage() | |||
| sys.exit() | |||
| if len(sys.argv[1:]) < 3: | |||
| usage() | |||
| sys.exit() | |||
| allDict = [] | |||
| for line in fileinput.input(inputFile): | |||
| m = re.match('^\#',line) | |||
| if m is not None: | |||
| pass | |||
| else: | |||
| oneDict = process(line) | |||
| allDict.append(oneDict) | |||
| allTable = pd.DataFrame(allDict) | |||
| allTable.to_csv(outputFile,sep='\t',index=False) | |||
+ 109
- 0
codescripts/oneClass.py
Dosyayı Görüntüle
| @@ -0,0 +1,109 @@ | |||
| # import modules | |||
| import numpy as np | |||
| import pandas as pd | |||
| from sklearn import svm | |||
| from sklearn import preprocessing | |||
| import sys, argparse, os | |||
| from vcf2bed import position_to_bed,padding_region | |||
| parser = argparse.ArgumentParser(description="this script is to preform one calss svm on each chromosome") | |||
| parser.add_argument('-train', '--trainDataset', type=str, help='training dataset generated from extracting vcf information part, with mutaitons supported by callsets', required=True) | |||
| parser.add_argument('-test', '--testDataset', type=str, help='testing dataset generated from extracting vcf information part, with mutaitons not called by all callsets', required=True) | |||
| parser.add_argument('-name', '--sampleName', type=str, help='sample name for output file name', required=True) | |||
| args = parser.parse_args() | |||
| # Rename input: | |||
| train_input = args.trainDataset | |||
| test_input = args.testDataset | |||
| sample_name = args.sampleName | |||
| # default columns, which will be included in the included in the calssifier | |||
| chromosome = ['chr1','chr2','chr3','chr4','chr5','chr6','chr7','chr8','chr9','chr10','chr11','chr12','chr13','chr14','chr15' ,'chr16','chr17','chr18','chr19','chr20','chr21','chr22','chrX','chrY'] | |||
| feature_heter_cols = ['AltDP','BaseQRankSum','DB','DP','FS','GQ','MQ','MQRankSum','QD','ReadPosRankSum','RefDP','SOR','af'] | |||
| feature_homo_cols = ['AltDP','DB','DP','FS','GQ','MQ','QD','RefDP','SOR','af'] | |||
| # import datasets sepearate the records with or without BaseQRankSum annotation, etc. | |||
| def load_dat(dat_file_name): | |||
| dat = pd.read_table(dat_file_name) | |||
| dat['DB'] = dat['DB'].fillna(0) | |||
| dat = dat[dat['DP'] != 0] | |||
| dat['af'] = dat['AltDP']/(dat['AltDP'] + dat['RefDP']) | |||
| homo_rows = dat[dat['BaseQRankSum'].isnull()] | |||
| heter_rows = dat[dat['BaseQRankSum'].notnull()] | |||
| return homo_rows,heter_rows | |||
| train_homo,train_heter = load_dat(train_input) | |||
| test_homo,test_heter = load_dat(test_input) | |||
| clf = svm.OneClassSVM(nu=0.05,kernel='rbf', gamma='auto_deprecated',cache_size=500) | |||
| def prepare_dat(train_dat,test_dat,feature_cols,chromo): | |||
| chr_train = train_dat[train_dat['chromo'] == chromo] | |||
| chr_test = test_dat[test_dat['chromo'] == chromo] | |||
| train_dat = chr_train.loc[:,feature_cols] | |||
| test_dat = chr_test.loc[:,feature_cols] | |||
| train_dat_scaled = preprocessing.scale(train_dat) | |||
| test_dat_scaled = preprocessing.scale(test_dat) | |||
| return chr_test,train_dat_scaled,test_dat_scaled | |||
| def oneclass(X_train,X_test,chr_test): | |||
| clf.fit(X_train) | |||
| y_pred_test = clf.predict(X_test) | |||
| test_true_dat = chr_test[y_pred_test == 1] | |||
| test_false_dat = chr_test[y_pred_test == -1] | |||
| return test_true_dat,test_false_dat | |||
| predicted_true = pd.DataFrame(columns=train_homo.columns) | |||
| predicted_false = pd.DataFrame(columns=train_homo.columns) | |||
| for chromo in chromosome: | |||
| # homo datasets | |||
| chr_test_homo,X_train_homo,X_test_homo = prepare_dat(train_homo,test_homo,feature_homo_cols,chromo) | |||
| test_true_homo,test_false_homo = oneclass(X_train_homo,X_test_homo,chr_test_homo) | |||
| predicted_true = predicted_true.append(test_true_homo) | |||
| predicted_false = predicted_false.append(test_false_homo) | |||
| # heter datasets | |||
| chr_test_heter,X_train_heter,X_test_heter = prepare_dat(train_heter,test_heter,feature_heter_cols,chromo) | |||
| test_true_heter,test_false_heter = oneclass(X_train_heter,X_test_heter,chr_test_heter) | |||
| predicted_true = predicted_true.append(test_true_heter) | |||
| predicted_false = predicted_false.append(test_false_heter) | |||
| predicted_true_filename = sample_name + '_predicted_true.txt' | |||
| predicted_false_filename = sample_name + '_predicted_false.txt' | |||
| predicted_true.to_csv(predicted_true_filename,sep='\t',index=False) | |||
| predicted_false.to_csv(predicted_false_filename,sep='\t',index=False) | |||
| # output the bed file and padding bed region 50bp | |||
| predicted_true_bed_filename = sample_name + '_predicted_true.bed' | |||
| predicted_false_bed_filename = sample_name + '_predicted_false.bed' | |||
| padding_filename = sample_name + '_padding.bed' | |||
| predicted_true_bed = open(predicted_true_bed_filename,'w') | |||
| predicted_false_bed = open(predicted_false_bed_filename,'w') | |||
| padding = open(padding_filename,'w') | |||
| # | |||
| for index,row in predicted_false.iterrows(): | |||
| chromo,pos1,pos2 = position_to_bed(row['chromo'],row['pos'],row['ref'],row['alt']) | |||
| outline_pos = chromo + '\t' + str(pos1) + '\t' + str(pos2) + '\n' | |||
| predicted_false_bed.write(outline_pos) | |||
| chromo,pad_pos1,pad_pos2,pad_pos3,pad_pos4 = padding_region(chromo,pos1,pos2,50) | |||
| outline_pad_1 = chromo + '\t' + str(pad_pos1) + '\t' + str(pad_pos2) + '\n' | |||
| outline_pad_2 = chromo + '\t' + str(pad_pos3) + '\t' + str(pad_pos4) + '\n' | |||
| padding.write(outline_pad_1) | |||
| padding.write(outline_pad_2) | |||
| for index,row in predicted_true.iterrows(): | |||
| chromo,pos1,pos2 = position_to_bed(row['chromo'],row['pos'],row['ref'],row['alt']) | |||
| outline_pos = chromo + '\t' + str(pos1) + '\t' + str(pos2) + '\n' | |||
| predicted_true_bed.write(outline_pos) | |||
+ 62
- 0
codescripts/select_small_variants_supported_by_all_callsets.py
Dosyayı Görüntüle
| @@ -0,0 +1,62 @@ | |||
| import sys,getopt | |||
| import os | |||
| import re | |||
| import fileinput | |||
| def usage(): | |||
| print( | |||
| """ | |||
| Usage: python select_small_variants_supported_by_all_callsets.py -i input_merged_vcf_file -o prefix | |||
| This script selects SNPs and Indels supported by all callsets. | |||
| Input: | |||
| -i a merged vcf file | |||
| Output: | |||
| -o a vcf file containd the selected SNPs and Indels | |||
| """) | |||
| # select supported small variants | |||
| def process(oneLine): | |||
| m = re.match('^\#',oneLine) | |||
| if m is not None: | |||
| outVCF.write(oneLine) | |||
| OUTname.write(oneLine) | |||
| else: | |||
| line = oneLine.rstrip() | |||
| strings = line.strip().split('\t') | |||
| gt = [i.split(':', 1)[0] for i in strings[9:len(strings)]] | |||
| if all(e == gt[0] for e in gt) and (gt[0] != '.'): | |||
| # output the record to vcf | |||
| outVCF.write(oneLine) | |||
| else: | |||
| OUTname.write(oneLine) | |||
| opts,args = getopt.getopt(sys.argv[1:],"hi:o:") | |||
| for op,value in opts: | |||
| if op == "-i": | |||
| inputFile=value | |||
| elif op == "-o": | |||
| prefix=value | |||
| elif op == "-h": | |||
| usage() | |||
| sys.exit() | |||
| if len(sys.argv[1:]) < 3: | |||
| usage() | |||
| sys.exit() | |||
| VCFname = prefix + '.vcf' | |||
| OUTname = prefix + '_outlier.vcf' | |||
| outVCF = open(VCFname,'w') | |||
| OUTname = open(OUTname,'w') | |||
| for line in fileinput.input(inputFile): | |||
| process(line) | |||
| outVCF.close() | |||
| OUTname.close() | |||
+ 36
- 0
codescripts/vcf2bed.py
Dosyayı Görüntüle
| @@ -0,0 +1,36 @@ | |||
| import re | |||
| def position_to_bed(chromo,pos,ref,alt): | |||
| # snv | |||
| # Start cooridinate BED = start coordinate VCF - 1 | |||
| # End cooridinate BED = start coordinate VCF | |||
| if len(ref) == 1 and len(alt) == 1: | |||
| StartPos = int(pos) -1 | |||
| EndPos = int(pos) | |||
| # deletions | |||
| # Start cooridinate BED = start coordinate VCF - 1 | |||
| # End cooridinate BED = start coordinate VCF + (reference length - alternate length) | |||
| elif len(ref) > 1 and len(alt) == 1: | |||
| StartPos = int(pos) - 1 | |||
| EndPos = int(pos) + (len(ref) - 1) | |||
| #insertions | |||
| # For insertions: | |||
| # Start cooridinate BED = start coordinate VCF - 1 | |||
| # End cooridinate BED = start coordinate VCF + (alternate length - reference length) | |||
| else: | |||
| StartPos = int(pos) - 1 | |||
| EndPos = int(pos) + (len(alt) - 1) | |||
| return chromo,StartPos,EndPos | |||
| def padding_region(chromo,pos1,pos2,padding): | |||
| StartPos1 = pos1 - padding | |||
| EndPos1 = pos1 | |||
| StartPos2 = pos2 | |||
| EndPos2 = pos2 + padding | |||
| return chromo,StartPos1,EndPos1,StartPos2,EndPos2 | |||
+ 33
- 0
family.tsv
Dosyayı Görüntüle
| @@ -0,0 +1,33 @@ | |||
| Quartet_DNA_BGI_SEQ2000_BGI_LCL5_1_20180518 oss://chinese-quartet/quartet-result-data/FDU/VCF/Quartet_DNA_BGI_SEQ2000_BGI_LCL5_1_20180518_hc.vcf | |||
| Quartet_DNA_BGI_SEQ2000_BGI_LCL5_2_20180530 oss://chinese-quartet/quartet-result-data/FDU/VCF/Quartet_DNA_BGI_SEQ2000_BGI_LCL5_2_20180530_hc.vcf | |||
| Quartet_DNA_BGI_SEQ2000_BGI_LCL5_3_20180530 oss://chinese-quartet/quartet-result-data/FDU/VCF/Quartet_DNA_BGI_SEQ2000_BGI_LCL5_3_20180530_hc.vcf | |||
| Quartet_DNA_BGI_SEQ2000_WGE_LCL5_1_20190402 oss://chinese-quartet/quartet-result-data/FDU/VCF/Quartet_DNA_BGI_SEQ2000_WGE_LCL5_1_20190402_hc.vcf | |||
| Quartet_DNA_BGI_SEQ2000_WGE_LCL5_2_20190402 oss://chinese-quartet/quartet-result-data/FDU/VCF/Quartet_DNA_BGI_SEQ2000_WGE_LCL5_2_20190402_hc.vcf | |||
| Quartet_DNA_BGI_SEQ500_BGI_LCL5_1_20180328 oss://chinese-quartet/quartet-result-data/FDU/VCF/Quartet_DNA_BGI_SEQ500_BGI_LCL5_1_20180328_hc.vcf | |||
| Quartet_DNA_BGI_SEQ500_BGI_LCL5_2_20180328 oss://chinese-quartet/quartet-result-data/FDU/VCF/Quartet_DNA_BGI_SEQ500_BGI_LCL5_2_20180328_hc.vcf | |||
| Quartet_DNA_BGI_SEQ500_BGI_LCL5_3_20180328 oss://chinese-quartet/quartet-result-data/FDU/VCF/Quartet_DNA_BGI_SEQ500_BGI_LCL5_3_20180328_hc.vcf | |||
| Quartet_DNA_ILM_Nova_ARD_LCL5_1_20181108 oss://chinese-quartet/quartet-result-data/FDU/VCF/Quartet_DNA_ILM_Nova_ARD_LCL5_1_20181108_RAW.vcf | |||
| Quartet_DNA_ILM_Nova_ARD_LCL5_2_20181108 oss://chinese-quartet/quartet-result-data/FDU/VCF/Quartet_DNA_ILM_Nova_ARD_LCL5_2_20181108_RAW.vcf | |||
| Quartet_DNA_ILM_Nova_ARD_LCL5_3_20181108 oss://chinese-quartet/quartet-result-data/FDU/VCF/Quartet_DNA_ILM_Nova_ARD_LCL5_3_20181108_RAW.vcf | |||
| Quartet_DNA_ILM_Nova_ARD_LCL5_4_20190111 oss://chinese-quartet/quartet-result-data/FDU/VCF/Quartet_DNA_ILM_Nova_ARD_LCL5_4_20190111_RAW.vcf | |||
| Quartet_DNA_ILM_Nova_ARD_LCL5_5_20190111 oss://chinese-quartet/quartet-result-data/FDU/VCF/Quartet_DNA_ILM_Nova_ARD_LCL5_5_20190111_RAW.vcf | |||
| Quartet_DNA_ILM_Nova_ARD_LCL5_6_20190111 oss://chinese-quartet/quartet-result-data/FDU/VCF/Quartet_DNA_ILM_Nova_ARD_LCL5_6_20190111_RAW.vcf | |||
| Quartet_DNA_ILM_Nova_BRG_LCL5_1_20171024 oss://chinese-quartet/quartet-result-data/FDU/VCF/Quartet_DNA_ILM_Nova_BRG_LCL5_1_20171024_RAW.vcf | |||
| Quartet_DNA_ILM_Nova_BRG_LCL5_1_20180930 oss://chinese-quartet/quartet-result-data/FDU/VCF/Quartet_DNA_ILM_Nova_BRG_LCL5_1_20180930_RAW.vcf | |||
| Quartet_DNA_ILM_Nova_BRG_LCL5_2_20180930 oss://chinese-quartet/quartet-result-data/FDU/VCF/Quartet_DNA_ILM_Nova_BRG_LCL5_2_20180930_RAW.vcf | |||
| Quartet_DNA_ILM_Nova_BRG_LCL5_3_20180930 oss://chinese-quartet/quartet-result-data/FDU/VCF/Quartet_DNA_ILM_Nova_BRG_LCL5_3_20180930_RAW.vcf | |||
| Quartet_DNA_ILM_Nova_GAC_LCL5_1_20171025 oss://chinese-quartet/quartet-result-data/FDU/VCF/Quartet_DNA_ILM_Nova_GAC_LCL5_1_20171025_RAW.vcf | |||
| Quartet_DNA_ILM_Nova_NVG_LCL5_1_20171024 oss://chinese-quartet/quartet-result-data/FDU/VCF/Quartet_DNA_ILM_Nova_NVG_LCL5_1_20171024_RAW.vcf | |||
| Quartet_DNA_ILM_Nova_WUX_LCL5_1_20171024 oss://chinese-quartet/quartet-result-data/FDU/VCF/Quartet_DNA_ILM_Nova_WUX_LCL5_1_20171024_RAW.vcf | |||
| Quartet_DNA_ILM_XTen_ARD_LCL5_1_20170403 oss://chinese-quartet/quartet-result-data/FDU/VCF/Quartet_DNA_ILM_XTen_ARD_LCL5_1_20170403_hc.vcf | |||
| Quartet_DNA_ILM_XTen_ARD_LCL5_2_20170403 oss://chinese-quartet/quartet-result-data/FDU/VCF/Quartet_DNA_ILM_XTen_ARD_LCL5_2_20170403_hc.vcf | |||
| Quartet_DNA_ILM_XTen_ARD_LCL5_3_20170403 oss://chinese-quartet/quartet-result-data/FDU/VCF/Quartet_DNA_ILM_XTen_ARD_LCL5_3_20170403_hc.vcf | |||
| Quartet_DNA_ILM_XTen_NVG_LCL5_1_20170329 oss://chinese-quartet/quartet-result-data/FDU/VCF/Quartet_DNA_ILM_XTen_NVG_LCL5_1_20170329_RAW.vcf | |||
| Quartet_DNA_ILM_XTen_NVG_LCL5_2_20170329 oss://chinese-quartet/quartet-result-data/FDU/VCF/Quartet_DNA_ILM_XTen_NVG_LCL5_2_20170329_RAW.vcf | |||
| Quartet_DNA_ILM_XTen_NVG_LCL5_3_20170329 oss://chinese-quartet/quartet-result-data/FDU/VCF/Quartet_DNA_ILM_XTen_NVG_LCL5_3_20170329_RAW.vcf | |||
| Quartet_DNA_ILM_XTen_WUX_LCL5_1_20170216 oss://chinese-quartet/quartet-result-data/FDU/VCF/Quartet_DNA_ILM_XTen_WUX_LCL5_1_20170216_RAW.vcf | |||
| Quartet_DNA_ILM_XTen_WUX_LCL5_2_20170216 oss://chinese-quartet/quartet-result-data/FDU/VCF/Quartet_DNA_ILM_XTen_WUX_LCL5_2_20170216_RAW.vcf | |||
| Quartet_DNA_ILM_XTen_WUX_LCL5_3_20170216 oss://chinese-quartet/quartet-result-data/FDU/VCF/Quartet_DNA_ILM_XTen_WUX_LCL5_3_20170216_RAW.vcf | |||
| Quartet_DNA_ILM_XTen_WUX_LCL5_4_20180703 oss://chinese-quartet/quartet-result-data/FDU/VCF/Quartet_DNA_ILM_XTen_WUX_LCL5_4_20180703_RAW.vcf | |||
| Quartet_DNA_ILM_XTen_WUX_LCL5_5_20180703 oss://chinese-quartet/quartet-result-data/FDU/VCF/Quartet_DNA_ILM_XTen_WUX_LCL5_5_20180703_RAW.vcf | |||
| Quartet_DNA_ILM_XTen_WUX_LCL5_6_20180703 oss://chinese-quartet/quartet-result-data/FDU/VCF/Quartet_DNA_ILM_XTen_WUX_LCL5_6_20180703_RAW.vcf | |||
+ 9
- 0
inputs
Dosyayı Görüntüle
| @@ -0,0 +1,9 @@ | |||
| { | |||
| "{{ project_name }}.mendelian.docker": "registry-vpc.cn-shanghai.aliyuncs.com/pgx-docker-registry/rtg-tools:latest", | |||
| "{{ project_name }}.disk_size": "100", | |||
| "{{ project_name }}.merge.docker": "registry-vpc.cn-shanghai.aliyuncs.com/pgx-docker-registry/rtg-tools:latest", | |||
| "{{ project_name }}.inputSamplesFile": "{{ inputSamplesFile }}", | |||
| "{{ project_name }}.sdf": "oss://chinese-quartet/quartet-storage-data/reference_data/GRCh38.d1.vd1.sdf/", | |||
| "{{ project_name }}.cluster_config": "OnDemand bcs.a2.large img-ubuntu-vpc", | |||
| "{{ project_name }}.KeepVar.docker": "registry-vpc.cn-shanghai.aliyuncs.com/pgx-docker-registry/high_confidence_call:v1.0" | |||
| } | |||
BIN
pictures/.DS_Store
Dosyayı Görüntüle
BIN
pictures/workflow.png
Dosyayı Görüntüle
BIN
tasks/.DS_Store
Dosyayı Görüntüle
+ 34
- 0
tasks/ExtractVCFinfo.wdl
Dosyayı Görüntüle
| @@ -0,0 +1,34 @@ | |||
| task ExtractVCFinfo { | |||
| File snv_train | |||
| File snv_test | |||
| File indel_train | |||
| File indel_test | |||
| String snv_train_sampleName = basename(snv_train,".vcf") | |||
| String snv_test_sampleName = basename(snv_test,".vcf") | |||
| String indel_train_sampleName = basename(indel_train,".vcf") | |||
| String indel_test_sampleName = basename(indel_test,".vcf") | |||
| String docker | |||
| String cluster_config | |||
| String disk_size | |||
| command <<< | |||
| python /opt/extract_vcf_information.py -i ${snv_train} -o ${snv_train_sampleName}.txt | |||
| python /opt/extract_vcf_information.py -i ${snv_test} -o ${snv_test_sampleName}.txt | |||
| python /opt/extract_vcf_information.py -i ${indel_train} -o ${indel_train_sampleName}.txt | |||
| python /opt/extract_vcf_information.py -i ${indel_test} -o ${indel_test_sampleName}.txt | |||
| >>> | |||
| runtime { | |||
| docker:docker | |||
| cluster: cluster_config | |||
| systemDisk: "cloud_ssd 40" | |||
| dataDisk: "cloud_ssd " + disk_size + " /cromwell_root/" | |||
| } | |||
| output { | |||
| File snv_train_vcf = "${snv_train_sampleName}.txt" | |||
| File snv_test_vcf = "${snv_test_sampleName}.txt" | |||
| File indel_train_vcf = "${indel_train_sampleName}.txt" | |||
| File indel_test_vcf = "${indel_test_sampleName}.txt" | |||
| } | |||
| } | |||
+ 26
- 0
tasks/KeepVar.wdl
Dosyayı Görüntüle
| @@ -0,0 +1,26 @@ | |||
| task KeepVar { | |||
| File violation_merged_vcf | |||
| File consistent_merged_vcf | |||
| String docker | |||
| String cluster_config | |||
| String disk_size | |||
| command <<< | |||
| python /opt/select_small_variants_supported_by_all_callsets.py -i ${violation_merged_vcf} -o violation.all.selected | |||
| python /opt/select_small_variants_supported_by_all_callsets.py -i ${consistent_merged_vcf} -o consistent.all.selected | |||
| >>> | |||
| runtime { | |||
| docker:docker | |||
| cluster: cluster_config | |||
| systemDisk: "cloud_ssd 40" | |||
| dataDisk: "cloud_ssd " + disk_size + " /cromwell_root/" | |||
| } | |||
| output { | |||
| File violation_keeped_vcf = "violation.all.selected.vcf" | |||
| File violation_outlier_vcf = "violation.all.selected_outlier.vcf" | |||
| File consistent_keeped_vcf = "consistent.all.selected.vcf" | |||
| File consistent_outlier_vcf = "consistent.all.selected_outlier.vcf" | |||
| } | |||
| } | |||
+ 69
- 0
tasks/SepSnvIndel.wdl
Dosyayı Görüntüle
| @@ -0,0 +1,69 @@ | |||
| task SepSnvIndel { | |||
| File vcf | |||
| String sampleName = basename(vcf,".normed.vcf") | |||
| File keeped_vcf | |||
| String docker | |||
| String cluster_config | |||
| String disk_size | |||
| command <<< | |||
| cat ${vcf} | grep '#' > header | |||
| cat ${vcf} | sed '/^#/d' | awk '$5!~/,/' > removed.body | |||
| cat ${vcf} | sed '/^#/d' | awk '$5~/,/' > MNP.body | |||
| cat header removed.body > ${sampleName}.MNPremoved.vcf | |||
| cat header MNP.body > ${sampleName}.MNP.vcf | |||
| rtg bgzip ${sampleName}.MNPremoved.vcf | |||
| rtg index -f vcf ${sampleName}.MNPremoved.vcf.gz | |||
| rtg bgzip ${keeped_vcf} -c > all.selected.vcf.gz | |||
| rtg index -f vcf all.selected.vcf.gz | |||
| rtg vcffilter -i ${sampleName}.MNPremoved.vcf.gz -o ${sampleName}.normed.snv.train.vcf.gz --include-vcf=all.selected.vcf.gz --snps-only | |||
| rtg vcffilter -i ${sampleName}.MNPremoved.vcf.gz -o ${sampleName}.normed.snv.test.vcf.gz --exclude-vcf=all.selected.vcf.gz --snps-only | |||
| rtg vcffilter -i ${sampleName}.MNPremoved.vcf.gz -o ${sampleName}.normed.indel.train.vcf.gz --include-vcf=all.selected.vcf.gz --non-snps-only | |||
| rtg vcffilter -i ${sampleName}.MNPremoved.vcf.gz -o ${sampleName}.normed.indel.test.vcf.gz --exclude-vcf=all.selected.vcf.gz --non-snps-only | |||
| rtg vcffilter -i ${sampleName}.MNPremoved.vcf.gz -o ${sampleName}.normed.snv.vcf.gz --snps-only | |||
| rtg vcffilter -i ${sampleName}.MNPremoved.vcf.gz -o ${sampleName}.normed.indel.vcf.gz --non-snps-only | |||
| gzip -d ${sampleName}.normed.snv.train.vcf.gz -c > ${sampleName}.normed.snv.train.vcf | |||
| gzip -d ${sampleName}.normed.snv.test.vcf.gz -c > ${sampleName}.normed.snv.test.vcf | |||
| gzip -d ${sampleName}.normed.indel.train.vcf.gz -c > ${sampleName}.normed.indel.train.vcf | |||
| gzip -d ${sampleName}.normed.indel.test.vcf.gz -c > ${sampleName}.normed.indel.test.vcf | |||
| >>> | |||
| runtime { | |||
| docker:docker | |||
| cluster: cluster_config | |||
| systemDisk: "cloud_ssd 40" | |||
| dataDisk: "cloud_ssd " + disk_size + " /cromwell_root/" | |||
| } | |||
| output { | |||
| File MNP="${sampleName}.MNP.vcf" | |||
| File snv_gz = "${sampleName}.normed.snv.vcf.gz" | |||
| File snv_idx = "${sampleName}.normed.snv.vcf.gz.tbi" | |||
| File indel_gz = "${sampleName}.normed.indel.vcf.gz" | |||
| File indel_idx = "${sampleName}.normed.indel.vcf.gz.tbi" | |||
| File snv_train = "${sampleName}.normed.snv.train.vcf" | |||
| File snv_test = "${sampleName}.normed.snv.test.vcf" | |||
| File indel_train = "${sampleName}.normed.indel.train.vcf" | |||
| File indel_test = "${sampleName}.normed.indel.test.vcf" | |||
| File snv_train_gz = "${sampleName}.normed.snv.train.vcf.gz" | |||
| File snv_test_gz = "${sampleName}.normed.snv.test.vcf.gz" | |||
| File indel_train_gz = "${sampleName}.normed.indel.train.vcf.gz" | |||
| File indel_test_gz = "${sampleName}.normed.indel.test.vcf.gz" | |||
| File snv_train_idx = "${sampleName}.normed.snv.train.vcf.gz.tbi" | |||
| File snv_test_idx = "${sampleName}.normed.snv.test.vcf.gz.tbi" | |||
| File indel_train_idx = "${sampleName}.normed.indel.train.vcf.gz.tbi" | |||
| File indel_test_idx = "${sampleName}.normed.indel.test.vcf.gz.tbi" | |||
| } | |||
| } | |||
+ 71
- 0
tasks/SepTrueFalse.wdl
Dosyayı Görüntüle
| @@ -0,0 +1,71 @@ | |||
| task SepTrueFalse { | |||
| File snv_true_bed | |||
| File snv_false_bed | |||
| File indel_true_bed | |||
| File indel_false_bed | |||
| File snv_padding | |||
| File indel_padding | |||
| File snv_gz | |||
| File indel_gz | |||
| File snv_idx | |||
| File indel_idx | |||
| File snv_test_gz | |||
| File indel_test_gz | |||
| File snv_test_idx | |||
| File indel_test_idx | |||
| String sampleName = basename(snv_gz,".normed.snv.vcf.gz") | |||
| String docker | |||
| String cluster_config | |||
| String disk_size | |||
| command <<< | |||
| rtg vcffilter -i ${snv_test_gz} -o ${sampleName}.true.snv.vcf.gz --include-bed=${snv_true_bed} | |||
| rtg vcffilter -i ${snv_test_gz} -o ${sampleName}.false.snv.vcf.gz --include-bed=${snv_false_bed} | |||
| rtg vcffilter -i ${snv_gz} -o ${sampleName}.remain.snv.vcf.gz --exclude-bed=${snv_false_bed} | |||
| rtg vcffilter -i ${snv_gz} -o ${sampleName}.padding.snv.vcf.gz --include-bed=${snv_padding} | |||
| rtg vcffilter -i ${indel_test_gz} -o ${sampleName}.true.indel.vcf.gz --include-bed=${indel_true_bed} | |||
| rtg vcffilter -i ${indel_test_gz} -o ${sampleName}.false.indel.vcf.gz --include-bed=${indel_false_bed} | |||
| rtg vcffilter -i ${indel_gz} -o ${sampleName}.remain.indel.vcf.gz --exclude-bed=${indel_false_bed} | |||
| rtg vcffilter -i ${indel_gz} -o ${sampleName}.padding.indel.vcf.gz --include-bed=${indel_padding} | |||
| >>> | |||
| runtime { | |||
| docker:docker | |||
| cluster: cluster_config | |||
| systemDisk: "cloud_ssd 40" | |||
| dataDisk: "cloud_ssd " + disk_size + " /cromwell_root/" | |||
| } | |||
| output { | |||
| File snv_true_vcf = "${sampleName}.true.snv.vcf.gz" | |||
| File snv_true_vcf_index = "${sampleName}.true.snv.vcf.gz.tbi" | |||
| File snv_false_vcf = "${sampleName}.false.snv.vcf.gz" | |||
| File snv_false_vcf_index = "${sampleName}.false.snv.vcf.gz.tbi" | |||
| File snv_remain_vcf = "${sampleName}.remain.snv.vcf.gz" | |||
| File snv_remain_vcf_index = "${sampleName}.remain.snv.vcf.gz.tbi" | |||
| File snv_padding_vcf = "${sampleName}.padding.snv.vcf.gz" | |||
| File snv_padding_vcf_index = "${sampleName}.padding.snv.vcf.gz.tbi" | |||
| File indel_true_vcf = "${sampleName}.true.indel.vcf.gz" | |||
| File indel_true_vcf_index = "${sampleName}.true.indel.vcf.gz.tbi" | |||
| File indel_false_vcf = "${sampleName}.false.indel.vcf.gz" | |||
| File indel_false_vcf_index = "${sampleName}.false.indel.vcf.gz.tbi" | |||
| File indel_remain_vcf = "${sampleName}.remain.indel.vcf.gz" | |||
| File indel_remain_vcf_index = "${sampleName}.remain.indel.vcf.gz.tbi" | |||
| File indel_padding_vcf = "${sampleName}.padding.indel.vcf.gz" | |||
| File indel_padding_vcf_index = "${sampleName}.padding.indel.vcf.gz.tbi" | |||
| } | |||
| } | |||
+ 29
- 0
tasks/indelNorm.wdl
Dosyayı Görüntüle
| @@ -0,0 +1,29 @@ | |||
| task indelNorm { | |||
| File vcf | |||
| File ref_dir | |||
| String fasta | |||
| String sampleName | |||
| String docker | |||
| String cluster_config | |||
| String disk_size | |||
| command <<< | |||
| cat ${vcf} | grep '#' > header | |||
| cat ${vcf} | grep -v '#' > body | |||
| cat body | grep -w '^chr1\|^chr2\|^chr3\|^chr4\|^chr5\|^chr6\|^chr7\|^chr8\|^chr9\|^chr10\|^chr11\|^chr12\|^chr13\|^chr14\|^chr15\|^chr16\|^chr17\|^chr18\|^chr19\|^chr20\|^chr21\|^chr22\|^chrX\|^chrY\|^chrM' > body.filtered | |||
| cat header body.filtered > ${sampleName}.filtered.vcf | |||
| /opt/hall-lab/bcftools-1.9/bin/bcftools norm -f ${ref_dir}/${fasta} ${sampleName}.filtered.vcf > ${sampleName}.normed.vcf | |||
| >>> | |||
| runtime { | |||
| docker:docker | |||
| cluster: cluster_config | |||
| systemDisk: "cloud_ssd 40" | |||
| dataDisk: "cloud_ssd " + disk_size + " /cromwell_root/" | |||
| } | |||
| output { | |||
| File normed_vcf = "${sampleName}.normed.vcf" | |||
| } | |||
| } | |||
+ 58
- 0
tasks/mendelian.wdl
Dosyayı Görüntüle
| @@ -0,0 +1,58 @@ | |||
| task mendelian { | |||
| File mother_vcf_gz | |||
| File father_vcf_gz | |||
| File child_vcf_gz | |||
| File mother_vcf_idx | |||
| File father_vcf_idx | |||
| File child_vcf_idx | |||
| String mother_name | |||
| String father_name | |||
| String child_name | |||
| String family_name | |||
| File sdf | |||
| String docker | |||
| String cluster_config | |||
| String disk_size | |||
| command <<< | |||
| rtg vcfmerge -o family.vcf.gz ${mother_vcf_gz} ${father_vcf_gz} ${child_vcf_gz} | |||
| rtg vcfannotate -i family.vcf.gz -o family.anno.vcf.gz \ | |||
| --add-header "##SAMPLE=<ID=${mother_name},Sex=FEMALE>" \ | |||
| --add-header "##SAMPLE=<ID=${father_name},Sex=MALE>" \ | |||
| --add-header "##SAMPLE=<ID=${child_name},Sex=FEMALE>" \ | |||
| --add-header "##PEDIGREE=<Child=${child_name},Mother=${mother_name},Father=${father_name}>" | |||
| rtg mendelian -i family.anno.vcf.gz -t ${sdf} -o ${family_name}.family.anno.mendelian.vcf.gz --lenient | |||
| zcat ${family_name}.family.anno.mendelian.vcf.gz | grep '#' > header | |||
| zcat ${family_name}.family.anno.mendelian.vcf.gz | grep -v '#'| grep 'MCU\|MCV' | cat header - > ${family_name}.violation.vcf | |||
| zcat ${family_name}.family.anno.mendelian.vcf.gz | grep -v '#'| grep -v 'MCV' | grep -v 'MCU' > ${family_name}.consistent.vcf | |||
| rtg bgzip ${family_name}.violation.vcf | |||
| rtg index -f vcf ${family_name}.violation.vcf.gz | |||
| rtg bgzip ${family_name}.consistent.vcf | |||
| rtg index -f vcf ${family_name}.consistent.vcf.gz | |||
| rtg vcffilter -i ${child_vcf_gz} -o ${child_name}.violation.vcf.gz --include-vcf=${family_name}.violation.vcf.gz | |||
| rtg vcffilter -i ${child_vcf_gz} -o ${child_name}.consistent.vcf.gz --include-vcf=${family_name}.consistent.vcf.gz | |||
| >>> | |||
| runtime { | |||
| docker:docker | |||
| cluster: cluster_config | |||
| systemDisk: "cloud_ssd 40" | |||
| dataDisk: "cloud_ssd " + disk_size + " /cromwell_root/" | |||
| } | |||
| output { | |||
| File family_vcf_gz = "${family_name}.family.anno.mendelian.vcf.gz" | |||
| File family_vcf_index = "${family_name}.family.anno.mendelian.vcf.gz.tbi" | |||
| File violation_vcf_gz = "${child_name}.violation.vcf.gz" | |||
| File violation_vcf_idx = "${child_name}.violation.vcf.gz.tbi" | |||
| File consistent_vcf_gz = "${child_name}.consistent.vcf.gz" | |||
| File consistent_vcf_idx = "${child_name}.consistent.vcf.gz.tbi" | |||
| } | |||
| } | |||
+ 26
- 0
tasks/merge.wdl
Dosyayı Görüntüle
| @@ -0,0 +1,26 @@ | |||
| task merge { | |||
| Array[File] violation_vcf_gz | |||
| Array[File] violation_vcf_idx | |||
| Array[File] consistent_vcf_gz | |||
| Array[File] consistent_vcf_idx | |||
| String docker | |||
| String cluster_config | |||
| String disk_size | |||
| command <<< | |||
| rtg vcfmerge --force-merge-all --no-gzip -o all.violation.vcf ${sep=" " violation_vcf_gz} | |||
| rtg vcfmerge --force-merge-all --no-gzip -o all.consistent.vcf ${sep=" " consistent_vcf_gz} | |||
| >>> | |||
| runtime { | |||
| docker:docker | |||
| cluster: cluster_config | |||
| systemDisk: "cloud_ssd 40" | |||
| dataDisk: "cloud_ssd " + disk_size + " /cromwell_root/" | |||
| } | |||
| output { | |||
| File violation_merged_vcf = "all.violation.vcf" | |||
| File consistent_merged_vcf = "all.consistent.vcf" | |||
| } | |||
| } | |||
+ 35
- 0
tasks/mergeBed.wdl
Dosyayı Görüntüle
| @@ -0,0 +1,35 @@ | |||
| task mergeBed { | |||
| Array[File] snv_true_bed | |||
| Array[File] snv_false_bed | |||
| Array[File] indel_true_bed | |||
| Array[File] indel_false_bed | |||
| Array[File] indel_padding | |||
| Array[File] snv_padding | |||
| String docker | |||
| String cluster_config | |||
| String disk_size | |||
| command <<< | |||
| /opt/ccdg/bedtools-2.27.1/bin/bedtools multiinter -i ${sep=" " snv_true_bed} ${sep=" " indel_true_bed} > merged.true.bed | |||
| /opt/ccdg/bedtools-2.27.1/bin/bedtools multiinter -i ${sep=" " snv_false_bed} ${sep=" " indel_false_bed} > merged.false.bed | |||
| /opt/ccdg/bedtools-2.27.1/bin/bedtools multiinter -i ${sep=" " snv_padding} ${sep=" " indel_padding} > merged.padding.bed | |||
| >>> | |||
| runtime { | |||
| docker:docker | |||
| cluster: cluster_config | |||
| systemDisk: "cloud_ssd 40" | |||
| dataDisk: "cloud_ssd " + disk_size + " /cromwell_root/" | |||
| } | |||
| output { | |||
| File true_bed = "merged.true.bed" | |||
| File false_bed = "merged.false.bed" | |||
| File padding = "merged.padding.bed" | |||
| } | |||
| } | |||
+ 61
- 0
tasks/mergeVCF.wdl
Dosyayı Görüntüle
| @@ -0,0 +1,61 @@ | |||
| task mergeVCF { | |||
| Array[File] snv_true_vcf | |||
| Array[File] snv_true_vcf_index | |||
| Array[File] snv_false_vcf | |||
| Array[File] snv_false_vcf_index | |||
| Array[File] snv_remain_vcf | |||
| Array[File] snv_remain_vcf_index | |||
| Array[File] snv_padding_vcf | |||
| Array[File] snv_padding_vcf_index | |||
| Array[File] indel_true_vcf | |||
| Array[File] indel_true_vcf_index | |||
| Array[File] indel_false_vcf | |||
| Array[File] indel_false_vcf_index | |||
| Array[File] indel_remain_vcf | |||
| Array[File] indel_remain_vcf_index | |||
| Array[File] indel_padding_vcf | |||
| Array[File] indel_padding_vcf_index | |||
| String quartet_sample | |||
| String docker | |||
| String cluster_config | |||
| String disk_size | |||
| command <<< | |||
| rtg vcfmerge --force-merge-all --no-gzip -o ${quartet_sample}.snv.true.vcf.gz ${sep=" " snv_true_vcf} | |||
| rtg vcfmerge --force-merge-all --no-gzip -o ${quartet_sample}.snv.false.vcf.gz ${sep=" " snv_false_vcf} | |||
| rtg vcfmerge --force-merge-all --no-gzip -o ${quartet_sample}.snv.remain.vcf.gz ${sep=" " snv_remain_vcf} | |||
| rtg vcfmerge --force-merge-all --no-gzip -o ${quartet_sample}.snv.padding.vcf.gz ${sep=" " snv_padding_vcf} | |||
| rtg vcfmerge --force-merge-all --no-gzip -o ${quartet_sample}.indel.true.vcf.gz ${sep=" " indel_true_vcf} | |||
| rtg vcfmerge --force-merge-all --no-gzip -o ${quartet_sample}.indel.false.vcf.gz ${sep=" " indel_false_vcf} | |||
| rtg vcfmerge --force-merge-all --no-gzip -o ${quartet_sample}.indel.remain.vcf.gz ${sep=" " indel_remain_vcf} | |||
| rtg vcfmerge --force-merge-all --no-gzip -o ${quartet_sample}.indel.padding.vcf.gz ${sep=" " indel_padding_vcf} | |||
| >>> | |||
| runtime { | |||
| docker:docker | |||
| cluster: cluster_config | |||
| systemDisk: "cloud_ssd 40" | |||
| dataDisk: "cloud_ssd " + disk_size + " /cromwell_root/" | |||
| } | |||
| output { | |||
| File merged_snv_true = "${quartet_sample}.snv.true.vcf.gz" | |||
| File merged_snv_false = "${quartet_sample}.snv.false.vcf.gz" | |||
| File merged_snv_remain = "${quartet_sample}.snv.remain.vcf.gz" | |||
| File merged_snv_padding = "${quartet_sample}.snv.padding.vcf.gz" | |||
| File merged_indel_true = "${quartet_sample}.indel.true.vcf.gz" | |||
| File merged_indel_false = "${quartet_sample}.indel.false.vcf.gz" | |||
| File merged_indel_remain = "${quartet_sample}.indel.remain.vcf.gz" | |||
| File merged_indel_padding = "${quartet_sample}.indel.padding.vcf.gz" | |||
| } | |||
| } | |||
+ 39
- 0
tasks/oneClass.wdl
Dosyayı Görüntüle
| @@ -0,0 +1,39 @@ | |||
| task oneClass { | |||
| File snv_train_vcf | |||
| File snv_test_vcf | |||
| File indel_train_vcf | |||
| File indel_test_vcf | |||
| String sampleName = basename(snv_train_vcf,".normed.snv.train.txt") | |||
| String docker | |||
| String cluster_config | |||
| String disk_size | |||
| command <<< | |||
| python /opt/oneClass.py -train ${snv_train_vcf} -test ${snv_test_vcf} -name ${sampleName}_snv | |||
| python /opt/oneClass.py -train ${indel_train_vcf} -test ${indel_test_vcf} -name ${sampleName}_indel | |||
| >>> | |||
| runtime { | |||
| docker:docker | |||
| cluster: cluster_config | |||
| systemDisk: "cloud_ssd 40" | |||
| dataDisk: "cloud_ssd " + disk_size + " /cromwell_root/" | |||
| } | |||
| output { | |||
| File snv_true_txt = "${sampleName}_snv_predicted_true.txt" | |||
| File snv_false_txt = "${sampleName}_snv_predicted_false.txt" | |||
| File snv_true_bed = "${sampleName}_snv_predicted_true.bed" | |||
| File snv_false_bed = "${sampleName}_snv_predicted_false.bed" | |||
| File snv_padding = "${sampleName}_snv_padding.bed" | |||
| File indel_true_txt = "${sampleName}_indel_predicted_true.txt" | |||
| File indel_false_txt = "${sampleName}_indel_predicted_false.txt" | |||
| File indel_true_bed = "${sampleName}_indel_predicted_true.bed" | |||
| File indel_false_bed = "${sampleName}_indel_predicted_false.bed" | |||
| File indel_padding = "${sampleName}_indel_padding.bed" | |||
| } | |||
| } | |||
+ 37
- 0
tasks/zipIndex.wdl
Dosyayı Görüntüle
| @@ -0,0 +1,37 @@ | |||
| task zipIndex { | |||
| File violation_vcf | |||
| File consistent_vcf | |||
| File child_vcf_gz | |||
| File child_vcf_idx | |||
| File child_name | |||
| String family_name | |||
| String docker | |||
| String cluster_config | |||
| String disk_size | |||
| command <<< | |||
| rtg bgzip ${violation_vcf} -c > ${family_name}.violation.vcf.gz | |||
| rtg index -f vcf ${family_name}.violation.vcf.gz | |||
| rtg bgzip ${consistent_vcf} -c > ${family_name}.consistent.vcf | |||
| rtg index -f vcf ${family_name}.consistent.vcf.gz | |||
| rtg vcffilter -i ${child_vcf_gz} -o ${child_name}.violation.vcf.gz --include-vcf=${family_name}.violation.vcf.gz | |||
| rtg vcffilter -i ${child_vcf_gz} -o ${child_name}.consistent.vcf.gz --include-vcf=${family_name}.consistent.vcf.gz | |||
| >>> | |||
| runtime { | |||
| docker:docker | |||
| cluster: cluster_config | |||
| systemDisk: "cloud_ssd 40" | |||
| dataDisk: "cloud_ssd " + disk_size + " /cromwell_root/" | |||
| } | |||
| output { | |||
| File violation_vcf_gz = "${child_name}.violation.vcf.gz" | |||
| File violation_vcf_idx = "${child_name}.violation.vcf.gz.tbi" | |||
| File consistent_vcf_gz = "${child_name}.consistent.vcf" | |||
| File consistent_vcf_idx = "${child_name}.consistent.vcf.tbi" | |||
| } | |||
| } | |||
+ 49
- 0
workflow.wdl
Dosyayı Görüntüle
| @@ -0,0 +1,49 @@ | |||
| import "./tasks/mendelian.wdl" as mendelian | |||
| import "./tasks/merge.wdl" as merge | |||
| import "./tasks/KeepVar.wdl" as KeepVar | |||
| workflow {{ project_name }} { | |||
| File inputSamplesFile | |||
| Array[Array[File]] inputSamples = read_tsv(inputSamplesFile) | |||
| File sdf | |||
| String cluster_config | |||
| String disk_size | |||
| scatter (sample in inputSamples){ | |||
| call mendelian.mendelian as mendelian { | |||
| input: | |||
| mother_vcf_gz=sample[0], | |||
| father_vcf_gz=sample[1], | |||
| child_vcf_gz=sample[2], | |||
| mother_vcf_idx=sample[3], | |||
| father_vcf_idx=sample[4], | |||
| child_vcf_idx=sample[5], | |||
| mother_name=sample[6], | |||
| father_name=sample[7], | |||
| child_name=sample[8], | |||
| family_name=sample[9], | |||
| sdf=sdf, | |||
| cluster_config=cluster_config, | |||
| disk_size=disk_size | |||
| } | |||
| } | |||
| call merge.merge as merge { | |||
| input: | |||
| violation_vcf_gz=mendelian.violation_vcf_gz, | |||
| violation_vcf_idx=mendelian.violation_vcf_idx, | |||
| consistent_vcf_gz=mendelian.consistent_vcf_gz, | |||
| consistent_vcf_idx=mendelian.consistent_vcf_idx, | |||
| cluster_config=cluster_config, | |||
| disk_size=disk_size | |||
| } | |||
| call KeepVar.KeepVar as KeepVar { | |||
| input: | |||
| violation_merged_vcf=merge.violation_merged_vcf, | |||
| consistent_merged_vcf=merge.consistent_merged_vcf, | |||
| cluster_config=cluster_config, | |||
| disk_size=disk_size | |||
| } | |||
| } | |||
Yükleniyor…