当前提交
e61f40c493
共有 6 个文件被更改,包括 151 次插入 和 0 次删除
+ 64
- 0
README.md
查看文件
| # BWA_DeepVariant | |||||
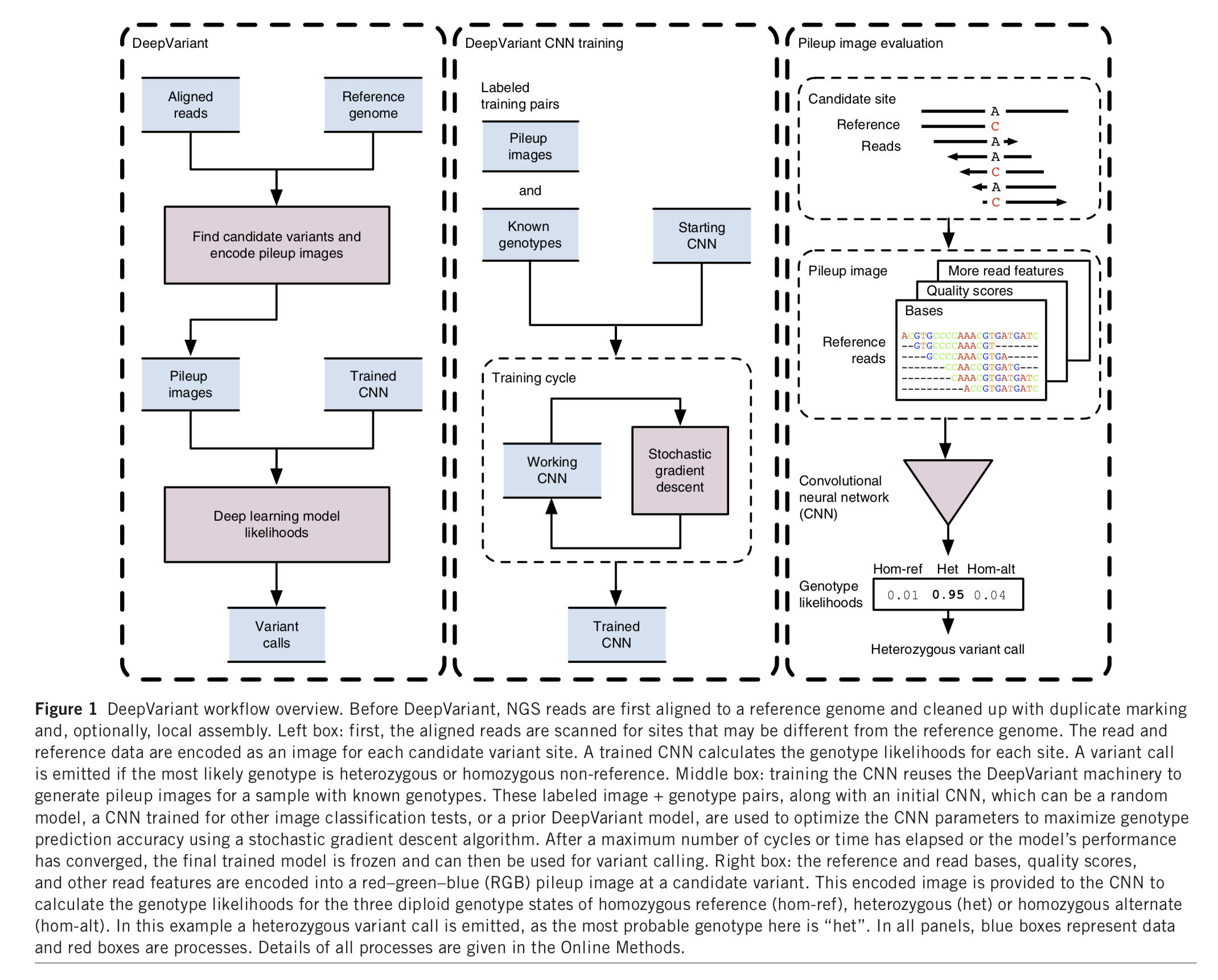
|  | |||||
| DeepVariant was trained on 8 whole genome replicates of NA12878 sequenced under a variety of conditions related to library preparation. These conditions include loading concentration, library size selection, and laboratory technician. Then the trained model can be generalized to a variety of new datasets and call variants. | |||||
| Docker uploaded to Alibaba Cloud is downloaded from [dockerhub](<https://hub.docker.com/r/dajunluo/deepvariant>) (version r0.8.0). | |||||
| WGS training datasets v0.8 include 12 HG001 PCR-free, 2 HG005 PCR-free, 4 HG001 PCR+. Sequencing platforms are all Illumina Hiseq. For Illumina Novaseq, BGISEQ-500 and BGISEQ-2000, we probably need to train customized small variants callers, more information can be found in [Github](<https://github.com/google/deepvariant/blob/r0.8/docs/deepvariant-tpu-training-case-study.md>). | |||||
| [CUSTOMIZED MODELS TO BE ADDED] | |||||
| DeepVariant pipeline consist of 3 steps: | |||||
| 1. `make_examples` consumes reads and the refernece genome to create TensorFlow examples for evaluation with deep learning models. | |||||
| 2. `call_variants` (Multiple-threads) consums TFRecord files of tf.Examples protos created by `make_examples` and a deep learning model checkpoint and evaluates the model on each example in the TFRecord. The output here is a TFRecord of CallVariantOutput protos. Multiple-threads | |||||
| 3. `postprocess_variants` (Single-thread) reads all of the output TFRecord files from `call_variants`, it needs to see all of the outputs from `call_variants`for a single sample to merge into a final VCF. | |||||
| **Some tips for DeepVariant:** | |||||
| 1. Duplicate marking may be performed, there is almost no difference inaccuracy except lower (<20x) coverages. | |||||
| 2. Authors recommend that you do not perform BQSR. Running BQSR has a small decrease on accuracy. | |||||
| 3. It is not necessary to do any form of indel realignment, though there is not a difference in DeepVariant accuracy either way. | |||||
| You can run with one common using the `run_deepvariant.py` script | |||||
| ```bash | |||||
| python run_deepvariant.py --model_type=WGS \ | |||||
| --ref=../../data/"${REFERENCE_FILE}" \ | |||||
| --reads=../../data/"${BAM_FILE}" \ | |||||
| --regions "chr20:10,000,000-10,010,000" \ | |||||
| --output_vcf=../output/output.vcf.gz \ | |||||
| --output_gvcf=../output/output.g.vcf.gz | |||||
| ``` | |||||
| Four files are generated | |||||
| ```bash | |||||
| output.vcf.gz | |||||
| output.vcf.gz.tbi | |||||
| output.g.vcf.gz | |||||
| output.g.vcf.gz.tbi | |||||
| ``` | |||||
| Command used in choppy app | |||||
| ```bas | |||||
| python run_deepvariant.py --model_type=WGS \ | |||||
| --ref=${ref_dir}/${fasta} \ | |||||
| --reads=${Dedup.bam} \ | |||||
| --output_vcf=${sample}_DP.vcf.gz | |||||
| ``` | |||||
| **Reference:** | |||||
| 1. DeepVariant Github <https://github.com/google/deepvariant> | |||||
| 2. DeepVariant paper <https://www.nature.com/articles/nbt.4235> | |||||
+ 11
- 0
defaults
查看文件
| { | |||||
| "Dedup_bam": "{{ Dedup_bam }}", | |||||
| "fasta": "GRCh38.d1.vd1.fa", | |||||
| "model_type": "WGS", | |||||
| "disk_size": "500", | |||||
| "DPdocker": "registry-vpc.cn-shanghai.aliyuncs.com/pgx-docker-registry/deepvariant:latest", | |||||
| "cluster_config": "OnDemand bcs.a2.7xlarge img-ubuntu-vpc", | |||||
| "Dedup_bam_index": "{{ Dedup_bam_index }}", | |||||
| "sample": "{{ sample }}", | |||||
| "ref_dir": "oss://pgx-reference-data/GRCh38.d1.vd1/" | |||||
| } |
+ 11
- 0
inputs
查看文件
| { | |||||
| "{{ project_name }}.Dedup_bam": "{{ Dedup_bam }}", | |||||
| "{{ project_name }}.fasta": "{{ fasta }}", | |||||
| "{{ project_name }}.model_type": "{{ model_type }}", | |||||
| "{{ project_name }}.disk_size": "{{ disk_size }}", | |||||
| "{{ project_name }}.DPdocker": "{{ DPdocker }}", | |||||
| "{{ project_name }}.cluster_config": "{{ cluster_config }}", | |||||
| "{{ project_name }}.Dedup_bam_index": "{{ Dedup_bam_index }}", | |||||
| "{{ project_name }}.sample": "{{ sample }}", | |||||
| "{{ project_name }}.ref_dir": "{{ ref_dir }}" | |||||
| } |
二进制
picture/Screen Shot 2019-06-11 at 10.09.27 AM.png
查看文件
+ 36
- 0
tasks/Deepvariant.wdl
查看文件
| task Deepvariant { | |||||
| File ref_dir | |||||
| String fasta | |||||
| File Dedup_bam | |||||
| File Dedup_bam_index | |||||
| String sample | |||||
| String model_type | |||||
| String DPdocker | |||||
| String cluster_config | |||||
| String disk_size | |||||
| command <<< | |||||
| set -o pipefail | |||||
| set -e | |||||
| nt=$(nproc) | |||||
| python /opt/deepvariant/bin/run_deepvariant.py --model_type=${model_type} \ | |||||
| --ref=${ref_dir}/${fasta} \ | |||||
| --reads=${Dedup_bam} \ | |||||
| --num_shards=$nt \ | |||||
| --call_variants_extra_args="use_openvino=true" \ | |||||
| --output_vcf=${sample}_DP.vcf.gz | |||||
| >>> | |||||
| runtime { | |||||
| docker:DPdocker | |||||
| cluster: cluster_config | |||||
| systemDisk: "cloud_ssd 40" | |||||
| dataDisk: "cloud_ssd " + disk_size + " /cromwell_root/" | |||||
| } | |||||
| output { | |||||
| File vcf = "${sample}_DP.vcf.gz" | |||||
| File vcf_index = "${sample}_DP.vcf.gz.tbi" | |||||
| } | |||||
| } |
+ 29
- 0
workflow.wdl
查看文件
| import "./tasks/Deepvariant.wdl" as Deepvariant | |||||
| workflow {{ project_name }} { | |||||
| File ref_dir | |||||
| String fasta | |||||
| File Dedup_bam | |||||
| File Dedup_bam_index | |||||
| String model_type | |||||
| String sample | |||||
| String DPdocker | |||||
| String cluster_config | |||||
| String disk_size | |||||
| call Deepvariant.Deepvariant as Deepvariant { | |||||
| input: | |||||
| fasta=fasta, | |||||
| ref_dir=ref_dir, | |||||
| Dedup_bam=Dedup_bam, | |||||
| model_type=model_type, | |||||
| Dedup_bam_index=Dedup_bam_index, | |||||
| sample=sample, | |||||
| DPdocker=DPdocker, | |||||
| cluster_config=cluster_config, | |||||
| disk_size=disk_size | |||||
| } | |||||
| } | |||||
正在加载...