當前提交
ca1163d323
共有 17 個檔案被更改,包括 1149 行新增 和 0 行删除
二進制
.DS_Store
查看文件
+ 356
- 0
README.md
查看文件
| @@ -0,0 +1,356 @@ | |||
| # RNA Sequencing Quality Control Pipeline | |||
| > Author: Li Zhihui | |||
| > | |||
| > E-mail:18210700119@fudan.edu.cn | |||
| > | |||
| > Git: http://choppy.3steps.cn/renluyao/RNAseq_germline_datapotal.git | |||
| > | |||
| > Last Updates: 2020/08/23 | |||
| ## 安装指南 | |||
| ``` | |||
| # 激活choppy环境 | |||
| source activate choppy | |||
| # 安装app | |||
| choppy install lizhihui/quartet-rnaseq-qc | |||
| ``` | |||
| ## App概述——中华家系1号标准物质介绍 | |||
| 建立高通量转录组测序的生物计量和质量控制关键技术体系,是保障转录组测序数据跨技术平台、跨实验室可比较、相关研究结果可重复、数据可共享的重要关键共性技术。建立国家转录组标准物质和基准数据集,突破基因组学的生物计量技术,是将测序技术转化成临床应用的重要环节与必经之路,目前国际上尚属空白。中国计量科学研究院与复旦大学、复旦大学泰州健康科学研究院共同研制了人源中华家系1号基因组标准物质(**Quartet,一套4个样本,编号分别为LCL5,LCL6,LCL7,LCL8,其中LCL5和LCL6为同卵双胞胎女儿,LCL7为父亲,LCL8为母亲**),以及相应的全基因组测序序列基准数据集(“量值”),为衡量基因序列检测准确与否提供一把“标尺”,成为保障基因测序数据可靠性的国家基准。人源中华家系1号转录组标准物质来源于泰州队列同卵双生双胞胎家庭,从遗传结构上体现了我国南北交界的人群结构特征,同时家系的设计也为“量值”的确定提供了遗传学依据。 | |||
| 中华家系1号RNA标准物质通过21个批次RNA标准物质的测序,整合集成了参考数据集,从而建立了一些依赖参考的质量控制指标。 | |||
| 在原始数据及比对数据层面,我们关注了数据量、GC含量、污染情况、比对率、比对位置分布等对测序可能会产生影响的指标,通过计算观察这些指标,我们可以一定程度上了解测序数据的质量。在测序数据表达情况层面,分别在定性和定量层面对基因检测表达水平进行了多角度的评价。通过从10个方面评估21个数据集的数据质量后我们构建了基于高置信度的已检测、未检测基因集、差异表达基因等参考数据集。通过这些指标,我们可以对新产生的测序数据进行合理的评价。 | |||
| 该Quality_control APP用于转录组测序(RNA Sequencing,RNA-Seq)数据的质量评估,包括原始数据及比对数据质控和基因表达数据质控。 | |||
| ## 流程与参数 | |||
| 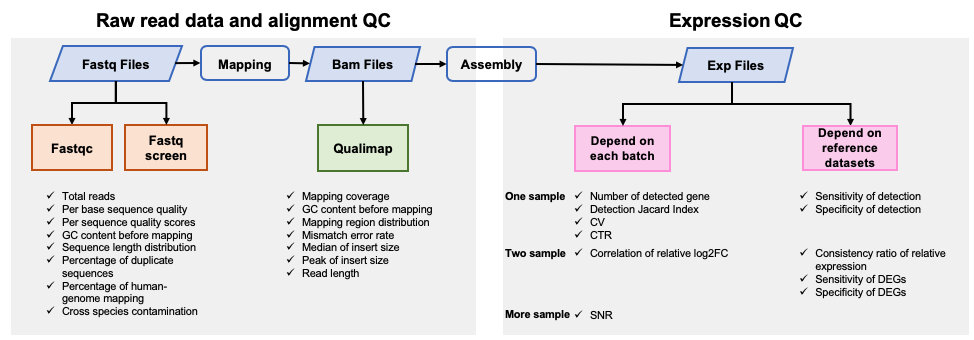 | |||
| ### 1.原始数据质量和数据比对质量 | |||
| #### [Fastqc](<https://www.bioinformatics.babraham.ac.uk/projects/fastqc/>) v0.11.5 | |||
| FastQC是一个常用的测序原始数据的质控软件,主要包括12个模块,具体请参考[Fastqc模块详情](<https://www.bioinformatics.babraham.ac.uk/projects/fastqc/Help/3%20Analysis%20Modules/>)。 | |||
| ```bash | |||
| fastqc -t <threads> -o <output_directory> <fastq_file> | |||
| ``` | |||
| #### [Fastq Screen](<https://www.bioinformatics.babraham.ac.uk/projects/fastq_screen/>) 0.12.0 | |||
| Fastq Screen是检测测序原始数据中是否引⼊入其他物种,或是接头引物等污染,⽐比如,如果测序样本 | |||
| 是⼈人类,我们期望99%以上的reads匹配到⼈人类基因组,10%左右的reads匹配到与⼈人类基因组同源性 | |||
| 较⾼高的⼩小⿏鼠上。如果有过多的reads匹配到Ecoli或者Yeast,要考虑是否在培养细胞的时候细胞系被污染,或者建库时⽂文库被污染。 | |||
| ```bash | |||
| fastq_screen --aligner <aligner> --conf <config_file> --top <number_of_reads> --threads <threads> <fastq_file> | |||
| ``` | |||
| `--conf` conifg 文件主要输入了多个物种的fasta文件地址,可根据自己自己的需求下载其他物种的fasta文件加入分析 | |||
| `--top`一般不需要对整个fastq文件进行检索,取前100000行 | |||
| #### [Qualimap](<http://qualimap.bioinfo.cipf.es/>) 2.0.0 | |||
| Qualimap是一个计算数据比对质量的软件,包含测序数据比对后的bam文件比对质量的结果。 | |||
| ```bash | |||
| qualimap bamqc -bam <bam_file> -outformat PDF:HTML -nt <threads> -outdir <output_directory> --java-mem-size=32G | |||
| qualimap rnaseq -bam ${bam} -outformat HTML -outdir ${bamname}_RNAseq -gtf ${gtf} -pe --java-mem-size=10G | |||
| ``` | |||
| ###2.数据表达质量 | |||
| ``` | |||
| Rscript | |||
| ``` | |||
| 分析采用实验室内部使用的代码,对从以下10个方面评估数据质量: | |||
| - Number of detected genes | |||
| - Detection Jaccard index (JI) | |||
| - Coefficient of variation (CV) | |||
| - Correlation of technical replicates (CTR) | |||
| - Sensitivity of detection | |||
| - Specificity of detection | |||
| - Consistency ratio of relative expression | |||
| - Correlation of relative log2FC | |||
| - Sensitivity of DEGs | |||
| - Specificity of DEGs | |||
| - Signal-to-noise Ratio (SNR) ) | |||
| ## App输入文件 | |||
| ``` | |||
| #read1 #read2 #sample_id #adapter_sequence #adapter_sequence_r2 | |||
| #待更新 | |||
| ``` | |||
| 参数设置: | |||
| 若有修改需求,请在input文件中添加新的行 | |||
| #### [fastp](https://github.com/OpenGene/fastp) | |||
| | 参数名 | 参数解释 | 默认值 | | |||
| | ------------------------- | ------------------------------------------------------- | ------------------------------------------------------------ | | |||
| | fastp_docker | fastp软件版本信息 | registry.cn-shanghai.aliyuncs.com/pgx-docker-registry/fastp:0.19.6 | | |||
| | fastp_cluster | fastp软件使用服务器 | OnDemand bcs.b2.3xlarge img-ubuntu-vpc | | |||
| | trim_front1 | 修剪read1前面多少个碱基 | 0 | | |||
| | trim_tail1 | 修剪read1尾部有多少个碱基 | 0 | | |||
| | max_len1 | 修剪read1的尾部使其与max_len1一样长 | 0 | | |||
| | trim_front2 | 修剪read2前面多少个碱基 | 0 | | |||
| | trim_tail2 | 修整read2尾部多少个碱基 | 0 | | |||
| | max_len2 | 修剪read2的尾部,使其与max_len2一样长 | 0 | | |||
| | adapter_sequence | R1端使用接头 | AGATCGGAAGAGCACACGTCTGAACTCCAGTCA | | |||
| | adapter_sequence_r2 | R2端使用接头 | AGATCGGAAGAGCGTCGTGTAGGGAAAGAGTGT | | |||
| | disable_adapter_trimming | 是否进行接头过滤(非0则不过滤) | 0 | | |||
| | length_required | 接头过滤参数:短于length_required的读取将被丢弃 | 50 | | |||
| | length_required1 | 接头过滤参数: 默认值20表示phred quality> = Q20是合格的 | 20 | | |||
| | UMI | 是否使用UMI接头(非0则使用) | 0 | | |||
| | umi_len | UMI接头参数: | 0 | | |||
| | umi_loc | UMI接头参数:接头位置 | umi_loc | | |||
| | disable_quality_filtering | 是否进行碱基质量过滤(非0则过滤) | 1 | | |||
| | qualified_quality_phred | 碱基质量过滤参数:允许不合格的百分比 | 20 | | |||
| #### [HISAT2](http://daehwankimlab.github.io/hisat2/) | |||
| | 参数名 | 参数解释 | 默认值 | | |||
| | ---------------- | ------------------------------------------------------------ | ------------------------------------------------------------ | | |||
| | hisat2_docker | hisat2软件版本信息 | registry.cn-shanghai.aliyuncs.com/pgx-docker-registry/hisat2:v2.1.0-2 | | |||
| | hisat2_cluster | hisat2软件使用服务器 | OnDemand | | |||
| | idx_prefix | 比对文件类型 | genome_snp_tran | | |||
| | idx | 比对文件地址 | oss://pgx-reference-data/reference/hisat2/grch38_snp_tran/ | | |||
| | fasta | 比对文件名称 | GRCh38.d1.vd1.fa | | |||
| | pen_cansplice | 为每对规范的剪接位点(例如GT / AG)设置惩罚 | 0 | | |||
| | pen_noncansplice | 设置每对非规范剪接位点(例如非GT / AG)的惩罚 | 3 | | |||
| | pen_intronlen | 设置长内含子的罚分,因此与较短的内含子相比,较短的内含子优先 | G,-8,1 | | |||
| | min_intronlen | 设置最小内含子长度 | 30 | | |||
| | max_intronlen | 设置最大内含子长度 | 500000 | | |||
| | maxins | 有效的配对末端比对的最大片段长度 | 500 | | |||
| | minins | 有效的配对末端比对的最小片段长度 | 0 | | |||
| ####[Samtools](http://www.htslib.org/) | |||
| | 参数名 | 参数解释 | 默认值 | | |||
| | ---------------- | ---------------------- | ------------------------------------------------------------ | | |||
| | samtools_docker | samtools软件版本信息 | registry.cn-shanghai.aliyuncs.com/pgx-docker-registry/samtools:v1.3.1 | | |||
| | samtools_cluster | samtools软件使用服务器 | OnDemand bcs.a2.large img-ubuntu-vpc, | | |||
| | insert_size | 最大插入读长 | 8000 | | |||
| ####[StringTie](https://ccb.jhu.edu/software/stringtie/) | |||
| | 参数名 | 参数解释 | 默认值 | | |||
| | ---------------------------------------------------- | ------------------------------------------------------------ | ------------------------------------------------------------ | | |||
| | stringtie_docker | stringtie软件版本信息 | registry.cn-shanghai.aliyuncs.com/pgx-docker-registry/stringtie:v1.3.4 | | |||
| | stringtie_cluster | stringtie软件使用服务器 | OnDemand bcs.a2.large img-ubuntu-vpc, | | |||
| | gtf | 组装gtf文件地址 | oss://pgx-reference-data/reference/annotation/Homo_sapiens.GRCh38.93.gtf | | |||
| | minimum_length_allowed_for_the_predicted_transcripts | 设置预测成绩单所允许的最小长度 | 200 | | |||
| | minimum_isoform_abundance | 将预测的转录本的最小同工型丰度设置为在给定基因座处组装的最丰富的转录本的一部分 | 0.01 | | |||
| | Junctions_no_spliced_reads | 没有拼接的接头在两端至少与该数量的碱基对齐,这些接头被过滤掉 | 10 | | |||
| | maximum_fraction_of_muliplelocationmapped_reads | 设置允许在给定基因座处存在的多核苷酸位置映射的读数的最大分数 | 0.95 | | |||
| #### [FastQC](https://www.bioinformatics.babraham.ac.uk/projects/fastqc/) | |||
| | 参数名 | 参数解释 | 默认值 | | |||
| | --------------------- | -------------------- | ------------------------------------------------------------ | | |||
| | fastqc_cluster_config | fastqc软件使用服务器 | OnDemand bcs.b2.3xlarge img-ubuntu-vpc | | |||
| | fastqc_docker | fastqc软件版本信息 | registry.cn-shanghai.aliyuncs.com/pgx-docker-registry/fastqc:v0.11.5 | | |||
| | fastqc_disk_size | fastqc文件盘大小 | 150 | | |||
| #### [Qualimap](http://qualimap.bioinfo.cipf.es/) | |||
| | 参数名 | 参数解释 | 默认值 | | |||
| | ----------------------------- | ---------------------------- | ------------------------------------------------------------ | | |||
| | qualimapBAMqc_docker | qualimapBAMqc软件版本信息 | registry.cn-shanghai.aliyuncs.com/pgx-docker-registry/qualimap:2.0.0 | | |||
| | qualimapBAMqc_cluster_config | qualimapBAMqc软件使用服务器 | OnDemand bcs.a2.7xlarge img-ubuntu-vpc | | |||
| | qualimapBAMqc_disk_size | qualimapBAMqc软件版本信息 | 500 | | |||
| | qualimapRNAseq_docker | qualimapRNAseq软件版本信息 | registry.cn-shanghai.aliyuncs.com/pgx-docker-registry/qualimap:2.0.0 | | |||
| | qualimapRNAseq_cluster_config | qualimapRNAseq软件使用服务器 | OnDemand bcs.a2.7xlarge img-ubuntu-vpc | | |||
| | qualimapRNAseq_disk_size | qualimapRNAseq软件版本信息 | 500 | | |||
| #### [FastQ Screen](https://www.bioinformatics.babraham.ac.uk/projects/fastq_screen/) | |||
| | 参数名 | 参数解释 | 默认值 | | |||
| | -------------------------- | --------------------------- | ------------------------------------------------------------ | | |||
| | fastqscreen_docker | fastqscreen软件版本信息 | registry.cn-shanghai.aliyuncs.com/pgx-docker-registry/fastqscreen:0.12.0 | | |||
| | fastqscreen_cluster_config | fastqscreen软件使用服务器 | OnDemand bcs.b2.3xlarge img-ubuntu-vpc | | |||
| | screen_ref_dir | fastqscreen软件序列地址 | oss://pgx-reference-data/fastq_screen_reference/ | | |||
| | fastq_screen_conf | fastqscreen软件序列索引地址 | oss://pgx-reference-data/fastq_screen_reference/fastq_screen.conf | | |||
| | fastqscreen_disk_size | fastqscreen文件盘大小 | 200 | | |||
| | ref_dir | fastqscreen序列索引地址 | oss://chinese-quartet/quartet-storage-data/reference_data/ | | |||
| #### [MultiQC](https://multiqc.info/) | |||
| | 参数名 | 参数解释 | 默认值 | | |||
| | ---------------------- | --------------------- | ------------------------------------------------------------ | | |||
| | multiqc_cluster_config | multiqc软件版本信息 | OnDemand bcs.b2.3xlarge img-ubuntu-vpc | | |||
| | multiqc_docker | multiqc软件使用服务器 | registry-vpc.cn-shanghai.aliyuncs.com/pgx-docker-registry/multiqc:v1.8 | | |||
| | multiqc_disk_size | multiqc文件盘大小 | 100 | | |||
| ## App输出文件 | |||
| #### 1. results_upstream_total.csv | |||
| | library | date | sample | replicate | Total.Sequences | GC_beforemapping | total_deduplicated_percentage | Human.percentage | ERCC.percentage | EColi.percentage | Adapter.percentage | Vector.percentage | rRNA.percentage | Virus.percentage | Yeast.percentage | Mitoch.percentage | Phix.percentage | No.hits.percentage | percentage_aligned_beforemapping | error_rate | bias_53 | GC_aftermapping | percent_duplicates | sequence_length | median_insert_size | mean_coverage | ins_size_median | ins_size_peak | exonic | intronic | intergenic | | |||
| | ------- | -------- | ------ | --------- | --------------- | ---------------- | ----------------------------- | ---------------- | --------------- | ---------------- | ------------------ | ----------------- | --------------- | ---------------- | ---------------- | ----------------- | --------------- | ------------------ | -------------------------------- | ---------- | ------- | --------------- | ------------------ | --------------- | ------------------ | ------------- | --------------- | ------------- | ------ | -------- | ---------- | | |||
| | D5_1 | 20200724 | D5 | 1 | 48872858 | 52 | 45.2953551 | 94.79 | 0 | 0 | 0.01 | 0.15 | 17.01 | 1.23 | 4.54 | 0.61 | 0 | 0.9 | 98.6435612 | 0.01 | 1.01 | 58.0426004 | 54.7046449 | 150 | 263 | 15.8021 | 258 | 192 | 52.05 | 41.37 | 6.58 | | |||
| | | | | | | | | | | | | | | | | | | | | | | | | | | | | | | | | | |||
| | | | | | | | | | | | | | | | | | | | | | | | | | | | | | | | | | |||
| 原始数据质量和数据比对质量结果汇总(example) | |||
| ### 2. One_sample.csv | |||
| | Name | Description | Group | Value | Reference_value | Conclusion | | |||
| | ------------------------ | ------------------------------------------------------------ | ----- | ---------- | --------------- | ---------- | | |||
| | Detected_gene | This metric is used to estimate the detection abundance of one sample. | D5 | 25126.6667 | (**, 58,395] | | | |||
| | | | D6 | 25858.6667 | (**, 58,395] | | | |||
| | | | F7 | 26089.6667 | (**, 58,395] | | | |||
| | | | M8 | 26618 | (**, 58,395] | | | |||
| | Jacard Index | Detection JI is the ratio of number of the genes detected in both replicates than the number of the genes detected in either of the replicates. This metric is used to estimate the repeatability of one sample detected gene from different replicates. | D5 | 0.8756 | [0.8, 1] | Pass | | |||
| | | | D6 | 0.8752 | [0.8, 1] | Pass | | |||
| | | | F7 | 0.8675 | [0.8, 1] | Pass | | |||
| | | | M8 | 0.8804 | [0.8, 1] | Pass | | |||
| | CV | CV is calculated based on the normalized expression levels in all 3 replicates of one sample for each genes. This metric is used to estimate the repeatability of one sample expression level from different replicates. | D5 | 11.4836 | | | | |||
| | | | D6 | 10.8401 | | | | |||
| | | | F7 | 12.2976 | | | | |||
| | | | M8 | 10.8662 | | | | |||
| | CTR | CTR is calculated based on the correlation of one sample expression level from different replicates. | D5 | 0.9718 | [0.95, 1] | Pass | | |||
| | | | D6 | 0.9737 | [0.95, 1] | Pass | | |||
| | | | F7 | 0.9699 | [0.95, 1] | Pass | | |||
| | | | M8 | 0.9725 | [0.95, 1] | Pass | | |||
| | Sensitivity_of_detection | Sensitivity is the proportion of true detected genes from reference dataset which can be correctly detected by the test set. | D5 | 0.9788 | [0.96, 1] | Pass | | |||
| | | | D6 | 0.9794 | [0.96, 1] | Pass | | |||
| | | | F7 | 0.9774 | [0.96, 1] | Pass | | |||
| | | | M8 | 0.9818 | [0.96, 1] | Pass | | |||
| | Specificity_of_detection | Specificity is the proportion of true non-detected genes from reference dataset which can be correctly not detected by the test set. | D5 | 0.9727 | [0.94, 1] | Pass | | |||
| | | | D6 | 0.9713 | [0.94, 1] | Pass | | |||
| | | | F7 | 0.9694 | [0.94, 1] | Pass | | |||
| | | | M8 | 0.9677 | [0.94, 1] | Pass | | |||
| 一个种类样本层面数据表达质量(example) | |||
| ### 3. Two_sample.csv | |||
| | Name | Description | Group | Value | Reference_value | Conclusion | | |||
| | ---------------------------------------- | ------------------------------------------------------------ | ----- | ---------- | --------------- | ---------- | | |||
| | Consistency_ratio_of_relative_expression | Proportion of genes that falls into reference range (mean +-2 fold SD) in relative ratio (log2FC). | D6/D5 | 1 | [0.82, 1] | Pass | | |||
| | | | F7/D5 | 1 | [0.82, 1] | Pass | | |||
| | | | F7/D6 | 1 | [0.82, 1] | Pass | | |||
| | | | M8/D5 | 1 | [0.82, 1] | Pass | | |||
| | | | M8/D6 | 1 | [0.82, 1] | Pass | | |||
| | | | M8/F7 | 1 | [0.82, 1] | Pass | | |||
| | Correlation_of_relative_log2FC | Pearson correlation between mean value of reference relative ratio and test site. | D6/D5 | 0.98137614 | [0.96,1] | Pass | | |||
| | | | F7/D5 | 0.9725557 | [0.96,1] | Pass | | |||
| | | | F7/D6 | 0.96789651 | [0.96,1] | Pass | | |||
| | | | M8/D5 | 0.97951286 | [0.96,1] | Pass | | |||
| | | | M8/D6 | 0.97959193 | [0.96,1] | Pass | | |||
| | | | M8/F7 | 0.97736629 | [0.96,1] | Pass | | |||
| | Sensitivity_of_DEGs | Sensitivity is the proportion of true DEGs from reference dataset which can be correctly identified as DEG by the test set. | D6/D5 | 0.8344293 | [0.80, 1] | Pass | | |||
| | | | F7/D5 | 0.84870451 | [0.80, 1] | Pass | | |||
| | | | F7/D6 | 0.84516486 | [0.80, 1] | Pass | | |||
| | | | M8/D5 | 0.86227581 | [0.80, 1] | Pass | | |||
| | | | M8/D6 | 0.86363942 | [0.80, 1] | Pass | | |||
| | | | M8/F7 | 0.85718483 | [0.80, 1] | Pass | | |||
| | Specificity_of_DEGs | Specificity is the proportion of true not DEGs from reference dataset which can be can be correctly identified as non-DEG by the test set. | D6/D5 | 0.97680659 | [0.95, 1] | Pass | | |||
| | | | F7/D5 | 0.97056775 | [0.95, 1] | Pass | | |||
| | | | F7/D6 | 0.975892 | [0.95, 1] | Pass | | |||
| | | | M8/D5 | 0.96896379 | [0.95, 1] | Pass | | |||
| | | | M8/D6 | 0.97206349 | [0.95, 1] | Pass | | |||
| | | | M8/F7 | 0.96594245 | [0.95, 1] | Pass | | |||
| 两个种类样本层面数据表达质量(example) | |||
| ### 4. More_sample.csv | |||
| | Name | Description | n | Value | Refenence_value | Conclusion | | |||
| | ---- | ------------------------------------------------------------ | ----- | ----- | --------------- | ---------- | | |||
| | SNR | Signal is defined as the average distance between libraries from the different samples on PCA plots and noise are those form the same samples. SNR is used to assess the ability to distinguish technical replicates from different biological samples. | 23705 | 13.64 | [5, inf) | Pass | | |||
| 多个种类样本层面数据表达质量(example) | |||
| ## 结果解读 | |||
| ### 1.原始数据质量和数据比对质量 | |||
| | 质控参数 | 软件 | 定义 | 参考值 | | |||
| | -------------------------------- | ------------ | ---------------------------- | --------- | | |||
| | Total.Sequences | Fastqc | 总读段数量 | > 10 M | | |||
| | GC_beforemapping | Fastqc | 比对前GC含量 | 40% - 60% | | |||
| | total_deduplicated_percentage | Fastqc | 重复序列比例 | | | |||
| | Human.percentage | FastQ Screen | 比对到人基因组读段比例 | > 90 % | | |||
| | ERCC.percentage | FastQ Screen | 比对到ERCC基因组读段比例 | < 5% | | |||
| | EColi.percentage | FastQ Screen | 比对到大肠杆菌基因组读段比例 | < 5% | | |||
| | Adapter.percentage | FastQ Screen | 比对到接头读段比例 | < 5% | | |||
| | Vector.percentage | FastQ Screen | 比对到载体读段比例 | < 5% | | |||
| | rRNA.percentage | FastQ Screen | 比对到接头读段比例 | < 10% | | |||
| | Virus.percentage | FastQ Screen | 比对到rRNA读段比例 | < 5% | | |||
| | Yeast.percentage | FastQ Screen | 比对到真菌读段比例 | < 5% | | |||
| | Mitoch.percentage | FastQ Screen | 比对到线粒体读段比例 | < 5% | | |||
| | Phix.percentage | FastQ Screen | 比对到Phix读段比例 | < 5% | | |||
| | No.hits.percentage | FastQ Screen | 未比对到已知物种读段比例 | < 5% | | |||
| | percentage_aligned_beforemapping | Qualimap | 比对率 | > 90% | | |||
| | error_rate | Qualimap | 错误率 | < 5% | | |||
| | bias_53 | Qualimap | 5-3偏好性 | | | |||
| | GC_aftermapping | Qualimap | 比对后GC含量 | 40% - 60% | | |||
| | percent_duplicates | Qualimap | 读段比对后重复比例 | | | |||
| | sequence_length | Qualimap | 读段长度 | ~150 | | |||
| | median_insert_size | Qualimap | 插入读段长度 | 200 - 300 | | |||
| | mean_coverage | Qualimap | 覆盖率 | | | |||
| | ins_size_median | Qualimap | 插入读段长度中位数 | 200 - 300 | | |||
| | ins_size_peak | Qualimap | 插入读段长度众数 | 200 - 300 | | |||
| | exonic | Qualimap | 比对到外显子的碱基比例 | 40% - 60% | | |||
| | intronic | Qualimap | 比对到内含子的碱基比例 | 40% - 60% | | |||
| | intergenic | Qualimap | 比对到内基因间区的碱基比例 | < 10% | | |||
| ###2.数据表达质量 | |||
| | Quality metrics | Category | Description | Reference value | | |||
| | ----------------------------------------- | ----------- | ------------------------------------------------------------ | --------------- | | |||
| | Number of detected genes | One group | This metric is used to estimate the detection abundance of one sample. | (**, 58,395] | | |||
| | Detection Jaccard index (JI) | One group | Detection JI is the ratio of number of the genes detected in both replicates than the number of the genes detected in either of the replicates. This metric is used to estimate the repeatability of one sample detected gene from different replicates. | [0.8, 1] | | |||
| | Coefficient of variation (CV) | One group | CV is calculated based on the normalized expression levels in all 3 replicates of one sample for each genes. This metric is used to estimate the repeatability of one sample expression level from different replicates. | [0, 0.2] | | |||
| | Correlation of technical replicates (CTR) | One group | CTR is calculated based on the correlation of one sample expression level from different replicates. | [0.95, 1] | | |||
| | Signal-to-noise Ratio (SNR) | More groups | Signal is defined as the average distance between libraries from the different samples on PCA plots and noise are those form the same samples. SNR is used to assess the ability to distinguish technical replicates from different biological samples. | [5, inf) | | |||
| | Sensitivity of detection | One group | Sensitivity is the proportion of "true" detected genes from reference dataset which can be correctly detected by the test set. | [0.96, 1] | | |||
| | Specificity of detection | One group | Specificity is the proportion of "true" non-detected genes from reference dataset which can be correctly not detected by the test set. | [0.94, 1] | | |||
| | Consistency ratio of relative expression | Two groups | Proportion of genes that falls into reference range (mean ± 2 fold SD) in relative ratio (log2FC). | [0.82, 1] | | |||
| | Correlation of relative log2FC | Two groups | Pearson correlation between mean value of reference relative ratio and test site. | [0.96,1] | | |||
| | Sensitivity of DEGs | Two groups | Sensitivity is the proportion of "true" DEGs from reference dataset which can be correctly identified as DEG by the test set. | [0.80, 1] | | |||
| | Specificity of DEGs | Two groups | Specificity is the proportion of "true" not DEGs from reference dataset which can be can be correctly identified as non-DEG by the test set. | [0.95, 1] | | |||
+ 94
- 0
conf/fastq_screen.conf
查看文件
| @@ -0,0 +1,94 @@ | |||
| # This is an example configuration file for FastQ Screen | |||
| ############################ | |||
| ## Bowtie, Bowtie 2 or BWA # | |||
| ############################ | |||
| ## If the Bowtie, Bowtie 2 or BWA binary is not in your PATH, you can set | |||
| ## this value to tell the program where to find your chosen aligner. Uncomment | |||
| ## the relevant line below and set the appropriate location. Please note, | |||
| ## this path should INCLUDE the executable filename. | |||
| #BOWTIE /usr/local/bin/bowtie/bowtie | |||
| #BOWTIE2 /usr/local/bowtie2/bowtie2 | |||
| #BWA /usr/local/bwa/bwa | |||
| ############################################ | |||
| ## Bismark (for bisulfite sequencing only) # | |||
| ############################################ | |||
| ## If the Bismark binary is not in your PATH then you can set this value to | |||
| ## tell the program where to find it. Uncomment the line below and set the | |||
| ## appropriate location. Please note, this path should INCLUDE the executable | |||
| ## filename. | |||
| #BISMARK /usr/local/bin/bismark/bismark | |||
| ############ | |||
| ## Threads # | |||
| ############ | |||
| ## Genome aligners can be made to run across multiple CPU cores to speed up | |||
| ## searches. Set this value to the number of cores you want for mapping reads. | |||
| THREADS 32 | |||
| ############## | |||
| ## DATABASES # | |||
| ############## | |||
| ## This section enables you to configure multiple genomes databases (aligner index | |||
| ## files) to search against in your screen. For each genome you need to provide a | |||
| ## database name (which can't contain spaces) and the location of the aligner index | |||
| ## files. | |||
| ## | |||
| ## The path to the index files SHOULD INCLUDE THE BASENAME of the index, e.g: | |||
| ## /data/public/Genomes/Human_Bowtie/GRCh37/Homo_sapiens.GRCh37 | |||
| ## Thus, the index files (Homo_sapiens.GRCh37.1.bt2, Homo_sapiens.GRCh37.2.bt2, etc.) | |||
| ## are found in a folder named 'GRCh37'. | |||
| ## | |||
| ## If, for example, the Bowtie, Bowtie2 and BWA indices of a given genome reside in | |||
| ## the SAME FOLDER, a SINLGE path may be provided to ALL the of indices. The index | |||
| ## used will be the one compatible with the chosen aligner (as specified using the | |||
| ## --aligner flag). | |||
| ## | |||
| ## The entries shown below are only suggested examples, you can add as many DATABASE | |||
| ## sections as required, and you can comment out or remove as many of the existing | |||
| ## entries as desired. We suggest including genomes and sequences that may be sources | |||
| ## of contamination either because they where run on your sequencer previously, or may | |||
| ## have contaminated your sample during the library preparation step. | |||
| ## | |||
| ## Human - sequences available from | |||
| ## ftp://ftp.ensembl.org/pub/current/fasta/homo_sapiens/dna/ | |||
| #DATABASE Human /data/public/Genomes/Human_Bowtie/GRCh37/Homo_sapiens.GRCh37 | |||
| ## | |||
| ## Mouse - sequence available from | |||
| ## ftp://ftp.ensembl.org/pub/current/fasta/mus_musculus/dna/ | |||
| #DATABASE Mouse /data/public/Genomes/Mouse/NCBIM37/Mus_musculus.NCBIM37 | |||
| ## | |||
| ## Ecoli- sequence available from EMBL accession U00096.2 | |||
| #DATABASE Ecoli /data/public/Genomes/Ecoli/Ecoli | |||
| ## | |||
| ## PhiX - sequence available from Refseq accession NC_001422.1 | |||
| #DATABASE PhiX /data/public/Genomes/PhiX/phi_plus_SNPs | |||
| ## | |||
| ## Adapters - sequence derived from the FastQC contaminats file found at: www.bioinformatics.babraham.ac.uk/projects/fastqc | |||
| #DATABASE Adapters /data/public/Genomes/Contaminants/Contaminants | |||
| ## | |||
| ## Vector - Sequence taken from the UniVec database | |||
| ## http://www.ncbi.nlm.nih.gov/VecScreen/UniVec.html | |||
| #DATABASE Vectors /data/public/Genomes/Vectors/Vectors | |||
| DATABASE Human /cromwell_root/tmp/fastq_screen_reference/genome | |||
| DATABASE Mouse /cromwell_root/tmp/fastq_screen_reference/mouse | |||
| DATABASE ERCC /cromwell_root/tmp/fastq_screen_reference/ERCC | |||
| DATABASE EColi /cromwell_root/tmp/fastq_screen_reference/ecoli | |||
| DATABASE Adapter /cromwell_root/tmp/fastq_screen_reference/adapters | |||
| DATABASE Vector /cromwell_root/tmp/fastq_screen_reference/vector | |||
| DATABASE rRNA /cromwell_root/tmp/fastq_screen_reference/rRNARef | |||
| DATABASE Virus /cromwell_root/tmp/fastq_screen_reference/viral | |||
| DATABASE Yeast /cromwell_root/tmp/fastq_screen_reference/GCF_000146045.2_R64_genomic_modify | |||
| DATABASE Mitoch /cromwell_root/tmp/fastq_screen_reference/Human_mitoch | |||
| DATABASE Phix /cromwell_root/tmp/fastq_screen_reference/phix | |||
+ 60
- 0
defaults
查看文件
| @@ -0,0 +1,60 @@ | |||
| { | |||
| "fastp_docker": "registry.cn-shanghai.aliyuncs.com/pgx-docker-registry/fastp:0.19.6", | |||
| "fastp_cluster": "OnDemand bcs.b2.3xlarge img-ubuntu-vpc", | |||
| "trim_front1": "0", | |||
| "trim_tail1": "0", | |||
| "max_len1": "0", | |||
| "trim_front2": "0", | |||
| "trim_tail2": "0", | |||
| "max_len2": "0", | |||
| "adapter_sequence": "AGATCGGAAGAGCACACGTCTGAACTCCAGTCA", | |||
| "adapter_sequence_r2": "AGATCGGAAGAGCGTCGTGTAGGGAAAGAGTGT", | |||
| "disable_adapter_trimming": "0", | |||
| "length_required": "50", | |||
| "length_required1": "20", | |||
| "UMI": "0", | |||
| "umi_len": "0", | |||
| "umi_loc": "umi_loc", | |||
| "qualified_quality_phred": "20", | |||
| "disable_quality_filtering": "1", | |||
| "hisat2_docker": "registry.cn-shanghai.aliyuncs.com/pgx-docker-registry/hisat2:v2.1.0-2", | |||
| "hisat2_cluster": "OnDemand bcs.a2.3xlarge img-ubuntu-vpc", | |||
| "idx_prefix": "genome_snp_tran", | |||
| "idx": "oss://pgx-reference-data/reference/hisat2/grch38_snp_tran/", | |||
| "pen_cansplice":"0", | |||
| "pen_noncansplice":"3", | |||
| "pen_intronlen":"G,-8,1", | |||
| "min_intronlen":"30", | |||
| "max_intronlen":"500000", | |||
| "maxins":"500", | |||
| "minins":"0", | |||
| "samtools_docker": "registry.cn-shanghai.aliyuncs.com/pgx-docker-registry/samtools:v1.3.1", | |||
| "samtools_cluster": "OnDemand bcs.a2.large img-ubuntu-vpc", | |||
| "insert_size":"8000", | |||
| "gtf": "oss://pgx-reference-data/reference/annotation/Homo_sapiens.GRCh38.93.gtf", | |||
| "stringtie_docker": "registry.cn-shanghai.aliyuncs.com/pgx-docker-registry/stringtie:v1.3.4", | |||
| "stringtie_cluster": "OnDemand bcs.a2.large img-ubuntu-vpc", | |||
| "minimum_length_allowed_for_the_predicted_transcripts":"200", | |||
| "minimum_isoform_abundance":"0.01", | |||
| "Junctions_no_spliced_reads":"10", | |||
| "maximum_fraction_of_muliplelocationmapped_reads":"0.95", | |||
| "fastqc_cluster_config": "OnDemand bcs.b2.3xlarge img-ubuntu-vpc", | |||
| "fastqc_docker": "registry.cn-shanghai.aliyuncs.com/pgx-docker-registry/fastqc:v0.11.5", | |||
| "fastqc_disk_size": "150", | |||
| "qualimapBAMqc_docker": "registry.cn-shanghai.aliyuncs.com/pgx-docker-registry/qualimap:2.0.0", | |||
| "qualimapBAMqc_cluster_config": "OnDemand bcs.a2.7xlarge img-ubuntu-vpc", | |||
| "qualimapBAMqc_disk_size": "500", | |||
| "qualimapRNAseq_docker": "registry.cn-shanghai.aliyuncs.com/pgx-docker-registry/qualimap:2.0.0", | |||
| "qualimapRNAseq_cluster_config": "OnDemand bcs.a2.7xlarge img-ubuntu-vpc", | |||
| "qualimapRNAseq_disk_size": "500", | |||
| "fastqscreen_docker": "registry.cn-shanghai.aliyuncs.com/pgx-docker-registry/fastqscreen:0.12.0", | |||
| "fastqscreen_cluster_config": "OnDemand bcs.b2.3xlarge img-ubuntu-vpc", | |||
| "screen_ref_dir": "oss://pgx-reference-data/fastq_screen_reference/", | |||
| "fastq_screen_conf": "oss://pgx-reference-data/fastq_screen_reference/fastq_screen.conf", | |||
| "fastqscreen_disk_size": "200", | |||
| "multiqc_cluster_config": "OnDemand bcs.b2.3xlarge img-ubuntu-vpc", | |||
| "multiqc_docker": "registry-vpc.cn-shanghai.aliyuncs.com/pgx-docker-registry/multiqc:v1.8", | |||
| "multiqc_disk_size": "100", | |||
| "ballgown_docker": "registry.cn-shanghai.aliyuncs.com/pgx-docker-registry/pgx-ballgown:0.0.1", | |||
| "ballgown_cluster": "OnDemand bcs.a2.large img-ubuntu-vpc" | |||
| } | |||
+ 66
- 0
inputs
查看文件
| @@ -0,0 +1,66 @@ | |||
| { | |||
| "{{ project_name }}.read1": "{{ read1 }}", | |||
| "{{ project_name }}.read2": "{{ read2 }}", | |||
| "{{ project_name }}.sample_id": "{{ sample_id }}", | |||
| "{{ project_name }}.fastp_docker": "{{ fastp_docker }}", | |||
| "{{ project_name }}.fastp_cluster": "{{ fastp_cluster }}", | |||
| "{{ project_name }}.trim_front1": "{{ trim_front1 }}", | |||
| "{{ project_name }}.trim_tail1": "{{ trim_tail1 }}", | |||
| "{{ project_name }}.max_len1": "{{ max_len1 }}", | |||
| "{{ project_name }}.trim_front2": "{{ trim_front2 }}", | |||
| "{{ project_name }}.trim_tail2": "{{ trim_tail2 }}", | |||
| "{{ project_name }}.max_len2": "{{ max_len2 }}", | |||
| "{{ project_name }}.adapter_sequence": "{{ adapter_sequence }}", | |||
| "{{ project_name }}.adapter_sequence_r2": "{{ adapter_sequence_r2 }}", | |||
| "{{ project_name }}.disable_adapter_trimming": "{{ disable_adapter_trimming }}", | |||
| "{{ project_name }}.length_required1": "{{ length_required1 }}", | |||
| "{{ project_name }}.UMI": "{{ UMI }}", | |||
| "{{ project_name }}.umi_loc": "{{ umi_loc }}", | |||
| "{{ project_name }}.umi_len": "{{ umi_len }}", | |||
| "{{ project_name }}.length_required": "{{ length_required }}", | |||
| "{{ project_name }}.qualified_quality_phred": "{{ qualified_quality_phred }}", | |||
| "{{ project_name }}.disable_quality_filtering": "{{ disable_quality_filtering }}", | |||
| "{{ project_name }}.hisat2_docker": "{{ hisat2_docker }}", | |||
| "{{ project_name }}.hisat2_cluster": "{{ hisat2_cluster }}", | |||
| "{{ project_name }}.idx_prefix": "{{ idx_prefix }}", | |||
| "{{ project_name }}.idx": "{{ idx }}", | |||
| "{{ project_name }}.fasta": "{{ fasta }}", | |||
| "{{ project_name }}.pen_cansplice": "{{ pen_cansplice }}", | |||
| "{{ project_name }}.pen_noncansplice": "{{ pen_noncansplice }}", | |||
| "{{ project_name }}.pen_intronlen": "{{ pen_intronlen }}", | |||
| "{{ project_name }}.min_intronlen": "{{ min_intronlen }}", | |||
| "{{ project_name }}.max_intronlen": "{{ max_intronlen }}", | |||
| "{{ project_name }}.maxins": "{{ maxins }}", | |||
| "{{ project_name }}.minins": "{{ minins }}", | |||
| "{{ project_name }}.samtools_docker": "{{ samtools_docker }}", | |||
| "{{ project_name }}.samtools_cluster": "{{ samtools_cluster }}", | |||
| "{{ project_name }}.insert_size": "{{ insert_size }}", | |||
| "{{ project_name }}.gtf": "{{ gtf }}", | |||
| "{{ project_name }}.stringtie_docker": "{{ stringtie_docker }}", | |||
| "{{ project_name }}.stringtie_cluster": "{{ stringtie_cluster }}", | |||
| "{{ project_name }}.minimum_length_allowed_for_the_predicted_transcripts": "{{ minimum_length_allowed_for_the_predicted_transcripts }}", | |||
| "{{ project_name }}.minimum_isoform_abundance": "{{ minimum_isoform_abundance }}", | |||
| "{{ project_name }}.Junctions_no_spliced_reads": "{{ Junctions_no_spliced_reads }}", | |||
| "{{ project_name }}.maximum_fraction_of_muliplelocationmapped_reads": "{{ maximum_fraction_of_muliplelocationmapped_reads }}", | |||
| "{{ project_name }}.fastqc_cluster_config": "{{ fastqc_cluster_config }}", | |||
| "{{ project_name }}.fastqc_docker": "{{ fastqc_docker }}", | |||
| "{{ project_name }}.fastqc_disk_size": "{{ fastqc_disk_size }}", | |||
| "{{ project_name }}.qualimapBAMqc_docker": "{{ qualimapBAMqc_docker }}", | |||
| "{{ project_name }}.qualimapBAMqc_cluster_config": "{{ qualimapBAMqc_cluster_config }}", | |||
| "{{ project_name }}.qualimapBAMqc_disk_size": "{{ qualimapBAMqc_disk_size }}", | |||
| "{{ project_name }}.qualimapRNAseq_docker": "{{ qualimapRNAseq_docker }}", | |||
| "{{ project_name }}.qualimapRNAseq_cluster_config": "{{ qualimapRNAseq_cluster_config }}", | |||
| "{{ project_name }}.qualimapRNAseq_disk_size": "{{ qualimapRNAseq_disk_size }}", | |||
| "{{ project_name }}.fastqscreen_docker": "{{ fastqscreen_docker }}", | |||
| "{{ project_name }}.fastqscreen_cluster_config": "{{ fastqscreen_cluster_config }}", | |||
| "{{ project_name }}.screen_ref_dir": "{{ screen_ref_dir }}", | |||
| "{{ project_name }}.fastq_screen_conf": "{{ fastq_screen_conf }}", | |||
| "{{ project_name }}.fastqscreen_disk_size": "{{ fastqscreen_disk_size }}", | |||
| "{{ project_name }}.ref_dir": "{{ ref_dir }}", | |||
| "{{ project_name }}.multiqc_cluster_config": "{{ multiqc_cluster_config }}", | |||
| "{{ project_name }}.multiqc_docker": "{{ multiqc_docker }}", | |||
| "{{ project_name }}.multiqc_disk_size": "{{ multiqc_disk_size }}", | |||
| "{{ project_name }}.ballgown_docker": "{{ ballgown_docker }}", | |||
| "{{ project_name }}.ballgown_cluster": "{{ ballgown_cluster }}", | |||
| "{{ project_name }}.disk_size": "{{ disk_size if disk_size != '' else 200}}" | |||
| } | |||
二進制
tasks/.DS_Store
查看文件
+ 25
- 0
tasks/ballgown.wdl
查看文件
| @@ -0,0 +1,25 @@ | |||
| task ballgown { | |||
| File gene_abundance | |||
| Array[File] ballgown | |||
| String sample_id | |||
| String docker | |||
| String cluster | |||
| String disk_size | |||
| command <<< | |||
| mkdir -p /cromwell_root/tmp/${sample_id} | |||
| cp -r ${sep=" " ballgown} /cromwell_root/tmp/${sample_id} | |||
| ballgown /cromwell_root/tmp/${sample_id} ${sample_id}.txt | |||
| >>> | |||
| runtime { | |||
| docker: docker | |||
| cluster: cluster | |||
| systemDisk: "cloud_ssd 40" | |||
| dataDisk: "cloud_ssd " + disk_size + " /cromwell_root/" | |||
| } | |||
| output { | |||
| File mat_expression = "${sample_id}.txt" | |||
| } | |||
| } | |||
+ 68
- 0
tasks/fastp.wdl
查看文件
| @@ -0,0 +1,68 @@ | |||
| task fastp { | |||
| String sample_id | |||
| File read1 | |||
| File read2 | |||
| String adapter_sequence | |||
| String adapter_sequence_r2 | |||
| String docker | |||
| String cluster | |||
| String disk_size | |||
| String umi_loc | |||
| Int trim_front1 | |||
| Int trim_tail1 | |||
| Int max_len1 | |||
| Int trim_front2 | |||
| Int trim_tail2 | |||
| Int max_len2 | |||
| Int disable_adapter_trimming | |||
| Int length_required | |||
| Int umi_len | |||
| Int UMI | |||
| Int qualified_quality_phred | |||
| Int length_required1 | |||
| Int disable_quality_filtering | |||
| command <<< | |||
| mkdir -p /cromwell_root/tmp/fastp/ | |||
| ##1.Disable_quality_filtering | |||
| if [ "${disable_quality_filtering}" == 0 ] | |||
| then | |||
| cp ${read1} /cromwell_root/tmp/fastp/{sample_id}_R1.fastq.tmp1.gz | |||
| cp ${read2} /cromwell_root/tmp/fastp/{sample_id}_R2.fastq.tmp1.gz | |||
| else | |||
| fastp --thread 4 --trim_front1 ${trim_front1} --trim_tail1 ${trim_tail1} --max_len1 ${max_len1} --trim_front2 ${trim_front2} --trim_tail2 ${trim_tail2} --max_len2 ${max_len2} -i ${read1} -I ${read2} -o /cromwell_root/tmp/fastp/${sample_id}_R1.fastq.tmp1.gz -O /cromwell_root/tmp/fastp/${sample_id}_R2.fastq.tmp1.gz -j ${sample_id}.json -h ${sample_id}.html | |||
| fi | |||
| ##2.UMI | |||
| if [ "${UMI}" == 0 ] | |||
| then | |||
| cp /cromwell_root/tmp/fastp/${sample_id}_R1.fastq.tmp1.gz /cromwell_root/tmp/fastp/${sample_id}_R1.fastq.tmp2.gz | |||
| cp /cromwell_root/tmp/fastp/${sample_id}_R2.fastq.tmp1.gz /cromwell_root/tmp/fastp/${sample_id}_R2.fastq.tmp2.gz | |||
| else | |||
| fastp --thread 4 -U --umi_loc=${umi_loc} --umi_len=${umi_len} --trim_front1 ${trim_front1} --trim_tail1 ${trim_tail1} --max_len1 ${max_len1} --trim_front2 ${trim_front2} --trim_tail2 ${trim_tail2} --max_len2 ${max_len2} -i /cromwell_root/tmp/fastp/${sample_id}_R1.fastq.tmp1.gz -I /cromwell_root/tmp/fastp/${sample_id}_R2.fastq.tmp1.gz -o /cromwell_root/tmp/fastp/${sample_id}_R1.fastq.tmp2.gz -O /cromwell_root/tmp/fastp/${sample_id}_R2.fastq.tmp2.gz -j ${sample_id}.json -h ${sample_id}.html | |||
| fi | |||
| ##3.Trim | |||
| if [ "${disable_adapter_trimming}" == 0 ] | |||
| then | |||
| fastp --thread 4 -l ${length_required} -q ${qualified_quality_phred} -u ${length_required1} --adapter_sequence ${adapter_sequence} --adapter_sequence_r2 ${adapter_sequence_r2} --detect_adapter_for_pe --trim_front1 ${trim_front1} --trim_tail1 ${trim_tail1} --max_len1 ${max_len1} --trim_front2 ${trim_front2} --trim_tail2 ${trim_tail2} --max_len2 ${max_len2} -i /cromwell_root/tmp/fastp/${sample_id}_R1.fastq.tmp2.gz -I /cromwell_root/tmp/fastp/${sample_id}_R2.fastq.tmp2.gz -o ${sample_id}_R1.fastq.gz -O ${sample_id}_R2.fastq.gz -j ${sample_id}.json -h ${sample_id}.html | |||
| else | |||
| cp /cromwell_root/tmp/fastp/${sample_id}_R1.fastq.tmp2.gz ${sample_id}_R1.fastq.gz | |||
| cp /cromwell_root/tmp/fastp/${sample_id}_R2.fastq.tmp2.gz ${sample_id}_R2.fastq.gz | |||
| fi | |||
| >>> | |||
| runtime { | |||
| docker: docker | |||
| cluster: cluster | |||
| systemDisk: "cloud_ssd 40" | |||
| dataDisk: "cloud_ssd " + disk_size + " /cromwell_root/" | |||
| } | |||
| output { | |||
| File json = "${sample_id}.json" | |||
| File report = "${sample_id}.html" | |||
| File Trim_R1 = "${sample_id}_R1.fastq.gz" | |||
| File Trim_R2 = "${sample_id}_R2.fastq.gz" | |||
| } | |||
| } | |||
+ 28
- 0
tasks/fastqc.wdl
查看文件
| @@ -0,0 +1,28 @@ | |||
| task fastqc { | |||
| File read1 | |||
| File read2 | |||
| String docker | |||
| String cluster_config | |||
| String disk_size | |||
| command <<< | |||
| set -o pipefail | |||
| set -e | |||
| nt=$(nproc) | |||
| fastqc -t $nt -o ./ ${read1} | |||
| fastqc -t $nt -o ./ ${read2} | |||
| >>> | |||
| runtime { | |||
| docker:docker | |||
| cluster: cluster_config | |||
| systemDisk: "cloud_ssd 40" | |||
| dataDisk: "cloud_ssd " + disk_size + " /cromwell_root/" | |||
| } | |||
| output { | |||
| File read1_html = sub(basename(read1), "\\.(fastq|fq)\\.gz$", "_fastqc.html") | |||
| File read1_zip = sub(basename(read1), "\\.(fastq|fq)\\.gz$", "_fastqc.zip") | |||
| File read2_html = sub(basename(read2), "\\.(fastq|fq)\\.gz$", "_fastqc.html") | |||
| File read2_zip = sub(basename(read2), "\\.(fastq|fq)\\.gz$", "_fastqc.zip") | |||
| } | |||
| } | |||
+ 37
- 0
tasks/fastqscreen.wdl
查看文件
| @@ -0,0 +1,37 @@ | |||
| task fastq_screen { | |||
| File read1 | |||
| File read2 | |||
| File screen_ref_dir | |||
| File fastq_screen_conf | |||
| String read1name = basename(read1,".fastq.gz") | |||
| String read2name = basename(read2,".fastq.gz") | |||
| String docker | |||
| String cluster_config | |||
| String disk_size | |||
| command <<< | |||
| set -o pipefail | |||
| set -e | |||
| nt=$(nproc) | |||
| mkdir -p /cromwell_root/tmp | |||
| cp -r ${screen_ref_dir} /cromwell_root/tmp/ | |||
| #sed -i "s#/cromwell_root/fastq_screen_reference#${screen_ref_dir}#g" ${fastq_screen_conf} | |||
| fastq_screen --aligner bowtie2 --conf ${fastq_screen_conf} --top 100000 --threads $nt ${read1} | |||
| fastq_screen --aligner bowtie2 --conf ${fastq_screen_conf} --top 100000 --threads $nt ${read2} | |||
| >>> | |||
| runtime { | |||
| docker:docker | |||
| cluster: cluster_config | |||
| systemDisk: "cloud_ssd 40" | |||
| dataDisk: "cloud_ssd " + disk_size + " /cromwell_root/" | |||
| } | |||
| output { | |||
| File png1 = "${read1name}_screen.png" | |||
| File txt1 = "${read1name}_screen.txt" | |||
| File html1 = "${read1name}_screen.html" | |||
| File png2 = "${read2name}_screen.png" | |||
| File txt2 = "${read2name}_screen.txt" | |||
| File html2 = "${read2name}_screen.html" | |||
| } | |||
| } | |||
+ 35
- 0
tasks/hisat2.wdl
查看文件
| @@ -0,0 +1,35 @@ | |||
| task hisat2 { | |||
| File idx | |||
| File Trim_R1 | |||
| File Trim_R2 | |||
| String idx_prefix | |||
| String sample_id | |||
| String docker | |||
| String cluster | |||
| String disk_size | |||
| String pen_intronlen | |||
| Int pen_cansplice | |||
| Int pen_noncansplice | |||
| Int min_intronlen | |||
| Int max_intronlen | |||
| Int maxins | |||
| Int minins | |||
| command <<< | |||
| nt=$(nproc) | |||
| hisat2 -t -p $nt -x ${idx}/${idx_prefix} --pen-cansplice ${pen_cansplice} --pen-noncansplice ${pen_noncansplice} --pen-intronlen ${pen_intronlen} --min-intronlen ${min_intronlen} --max-intronlen ${max_intronlen} --maxins ${maxins} --minins ${minins} --un-conc-gz ${sample_id}_un.fq.gz -1 ${Trim_R1} -2 ${Trim_R2} -S ${sample_id}.sam | |||
| >>> | |||
| runtime { | |||
| docker: docker | |||
| cluster: cluster | |||
| systemDisk: "cloud_ssd 40" | |||
| dataDisk: "cloud_ssd " + disk_size + " /cromwell_root/" | |||
| } | |||
| output { | |||
| File sam = "${sample_id}.sam" | |||
| File unmapread_1p = "${sample_id}_un.fq.1.gz" | |||
| File unmapread_2p = "${sample_id}_un.fq.2.gz" | |||
| } | |||
| } | |||
+ 61
- 0
tasks/multiqc.wdl
查看文件
| @@ -0,0 +1,61 @@ | |||
| task multiqc { | |||
| Array[File] read1_zip | |||
| Array[File] read2_zip | |||
| Array[File] txt1 | |||
| Array[File] txt2 | |||
| Array[File] bamqc_zip | |||
| Array[File] rnaseq_zip | |||
| String docker | |||
| String cluster_config | |||
| String disk_size | |||
| command <<< | |||
| set -o pipefail | |||
| set -e | |||
| mkdir -p /cromwell_root/tmp/fastqc | |||
| mkdir -p /cromwell_root/tmp/fastqscreen | |||
| mkdir -p /cromwell_root/tmp/bamqc | |||
| mkdir -p /cromwell_root/tmp/rnaseq | |||
| cp ${sep=" " read1_zip} ${sep=" " read2_zip} /cromwell_root/tmp/fastqc | |||
| cp ${sep=" " txt1} ${sep=" " txt2} /cromwell_root/tmp/fastqscreen | |||
| for i in ${sep=" " bamqc_zip} | |||
| do | |||
| tar -zxvf $i -C /cromwell_root/tmp/bamqc | |||
| done | |||
| for i in ${sep=" " rnaseq_zip} | |||
| do | |||
| tar -zxvf $i -C /cromwell_root/tmp/rnaseq | |||
| done | |||
| multiqc /cromwell_root/tmp/ | |||
| cat multiqc_data/multiqc_fastq_screen.txt > multiqc_fastq_screen.txt | |||
| cat multiqc_data/multiqc_fastqc.txt > multiqc_fastqc.txt | |||
| cat multiqc_data/multiqc_general_stats.txt > multiqc_general_stats.txt | |||
| cat multiqc_data/multiqc_qualimap_bamqc_genome_results.txt > multiqc_qualimap_bamqc_genome_results.txt | |||
| >>> | |||
| runtime { | |||
| docker:docker | |||
| cluster:cluster_config | |||
| systemDisk:"cloud_ssd 40" | |||
| dataDisk:"cloud_ssd " + disk_size + " /cromwell_root/" | |||
| } | |||
| output { | |||
| File multiqc_html = "multiqc_report.html" | |||
| Array[File] multiqc_txt = glob("multiqc_data/*") | |||
| File multiqc_fastq_screen = "multiqc_fastq_screen.txt" | |||
| File multiqc_fastqc = "multiqc_fastqc.txt" | |||
| File multiqc_general_stats = "multiqc_general_stats.txt" | |||
| File bamqc_genome_results = "multiqc_qualimap_bamqc_genome_results.txt" | |||
| } | |||
| } | |||
+ 28
- 0
tasks/qualimapBAMqc.wdl
查看文件
| @@ -0,0 +1,28 @@ | |||
| task qualimapBAMqc { | |||
| File bam | |||
| String bamname = basename(bam,".bam") | |||
| String docker | |||
| String cluster_config | |||
| String disk_size | |||
| command <<< | |||
| set -o pipefail | |||
| set -e | |||
| nt=$(nproc) | |||
| /opt/qualimap/qualimap bamqc -bam ${bam} -outformat PDF:HTML -nt $nt -outdir ${bamname}_bamqc --java-mem-size=32G | |||
| tar -zcvf ${bamname}_bamqc_qualimap.zip ${bamname}_bamqc | |||
| >>> | |||
| runtime { | |||
| docker:docker | |||
| cluster:cluster_config | |||
| systemDisk:"cloud_ssd 40" | |||
| dataDisk:"cloud_ssd " + disk_size + " /cromwell_root/" | |||
| } | |||
| output { | |||
| File bamqc_zip = "${bamname}_bamqc_qualimap.zip" | |||
| } | |||
| } | |||
+ 29
- 0
tasks/qualimapRNAseq.wdl
查看文件
| @@ -0,0 +1,29 @@ | |||
| task qualimapRNAseq { | |||
| File bam | |||
| File gtf | |||
| String bamname = basename(bam,".bam") | |||
| String docker | |||
| String cluster_config | |||
| String disk_size | |||
| command <<< | |||
| set -o pipefail | |||
| set -e | |||
| nt=$(nproc) | |||
| /opt/qualimap/qualimap rnaseq -bam ${bam} -outformat HTML -outdir ${bamname}_RNAseq -gtf ${gtf} -pe --java-mem-size=10G | |||
| tar -zcvf ${bamname}_RNAseq_qualimap.zip ${bamname}_RNAseq | |||
| >>> | |||
| runtime { | |||
| docker:docker | |||
| cluster:cluster_config | |||
| systemDisk:"cloud_ssd 40" | |||
| dataDisk:"cloud_ssd " + disk_size + " /cromwell_root/" | |||
| } | |||
| output { | |||
| File rnaseq_zip = "${bamname}_RNAseq_qualimap.zip" | |||
| } | |||
| } | |||
+ 39
- 0
tasks/samtools.wdl
查看文件
| @@ -0,0 +1,39 @@ | |||
| task samtools { | |||
| File sam | |||
| String sample_id | |||
| String bam = sample_id + ".bam" | |||
| String sorted_bam = sample_id + ".sorted.bam" | |||
| String percent_bam = sample_id + ".percent.bam" | |||
| String sorted_bam_index = sample_id + ".sorted.bam.bai" | |||
| String ins_size = sample_id + ".ins_size" | |||
| String docker | |||
| String cluster | |||
| String disk_size | |||
| Int insert_size | |||
| command <<< | |||
| set -o pipefail | |||
| set -e | |||
| /opt/conda/bin/samtools view -bS ${sam} > ${bam} | |||
| /opt/conda/bin/samtools sort -m 1000000000 ${bam} -o ${sorted_bam} | |||
| /opt/conda/bin/samtools index ${sorted_bam} | |||
| /opt/conda/bin/samtools view -bs 42.1 ${sorted_bam} > ${percent_bam} | |||
| /opt/conda/bin/samtools stats -i ${insert_size} ${sorted_bam} |grep ^IS|cut -f 2- > ${sample_id}.ins_size | |||
| >>> | |||
| runtime { | |||
| docker: docker | |||
| cluster: cluster | |||
| systemDisk: "cloud_ssd 40" | |||
| dataDisk: "cloud_ssd " + disk_size + " /cromwell_root/" | |||
| } | |||
| output { | |||
| File out_bam = sorted_bam | |||
| File out_percent = percent_bam | |||
| File out_bam_index = sorted_bam_index | |||
| File out_ins_size = ins_size | |||
| } | |||
| } | |||
+ 33
- 0
tasks/stringtie.wdl
查看文件
| @@ -0,0 +1,33 @@ | |||
| task stringtie { | |||
| File bam | |||
| File gtf | |||
| String docker | |||
| String sample_id | |||
| String cluster | |||
| String disk_size | |||
| Int minimum_length_allowed_for_the_predicted_transcripts | |||
| Int Junctions_no_spliced_reads | |||
| Float minimum_isoform_abundance | |||
| Float maximum_fraction_of_muliplelocationmapped_reads | |||
| command <<< | |||
| nt=$(nproc) | |||
| mkdir ballgown | |||
| /opt/conda/bin/stringtie -e -B -p $nt -f ${minimum_isoform_abundance} -m ${minimum_length_allowed_for_the_predicted_transcripts} -a ${Junctions_no_spliced_reads} -M ${maximum_fraction_of_muliplelocationmapped_reads} -G ${gtf} -o ballgown/${sample_id}/${sample_id}.gtf -C ${sample_id}.cov.ref.gtf -A ${sample_id}.gene.abundance.txt ${bam} | |||
| >>> | |||
| runtime { | |||
| docker: docker | |||
| cluster: cluster | |||
| systemDisk: "cloud_ssd 40" | |||
| dataDisk: "cloud_ssd " + disk_size + " /cromwell_root/" | |||
| } | |||
| output { | |||
| File covered_transcripts = "${sample_id}.cov.ref.gtf" | |||
| File gene_abundance = "${sample_id}.gene.abundance.txt" | |||
| Array[File] ballgown = ["ballgown/${sample_id}/${sample_id}.gtf", "ballgown/${sample_id}/e2t.ctab", "ballgown/${sample_id}/e_data.ctab", "ballgown/${sample_id}/i2t.ctab", "ballgown/${sample_id}/i_data.ctab", "ballgown/${sample_id}/t_data.ctab"] | |||
| File genecount = "{sample_id}_genecount.csv" | |||
| } | |||
| } | |||
+ 190
- 0
workflow.wdl
查看文件
| @@ -0,0 +1,190 @@ | |||
| import "./tasks/fastp.wdl" as fastp | |||
| import "./tasks/hisat2.wdl" as hisat2 | |||
| import "./tasks/samtools.wdl" as samtools | |||
| import "./tasks/stringtie.wdl" as stringtie | |||
| import "./tasks/fastqc.wdl" as fastqc | |||
| import "./tasks/fastqscreen.wdl" as fastqscreen | |||
| import "./tasks/qualimapBAMqc.wdl" as qualimapBAMqc | |||
| import "./tasks/qualimapRNAseq.wdl" as qualimapRNAseq | |||
| import "./tasks/ballgown.wdl" as ballgown | |||
| workflow {{ project_name }} { | |||
| File read1 | |||
| File read2 | |||
| File idx | |||
| File screen_ref_dir | |||
| File fastq_screen_conf | |||
| File gtf | |||
| String sample_id | |||
| String fastp_docker | |||
| String adapter_sequence | |||
| String adapter_sequence_r2 | |||
| String fastp_cluster | |||
| String umi_loc | |||
| String idx_prefix | |||
| String pen_intronlen | |||
| String fastqc_cluster_config | |||
| String fastqc_docker | |||
| String fastqscreen_docker | |||
| String fastqscreen_cluster_config | |||
| String hisat2_docker | |||
| String hisat2_cluster | |||
| String qualimapBAMqc_docker | |||
| String qualimapBAMqc_cluster_config | |||
| String qualimapRNAseq_docker | |||
| String qualimapRNAseq_cluster_config | |||
| String samtools_docker | |||
| String samtools_cluster | |||
| String stringtie_docker | |||
| String stringtie_cluster | |||
| String multiqc_cluster_config | |||
| String multiqc_docker | |||
| Int multiqc_disk_size | |||
| Int trim_front1 | |||
| Int trim_tail1 | |||
| Int max_len1 | |||
| Int trim_front2 | |||
| Int trim_tail2 | |||
| Int max_len2 | |||
| Int disable_adapter_trimming | |||
| Int length_required | |||
| Int umi_len | |||
| Int UMI | |||
| Int qualified_quality_phred | |||
| Int length_required1 | |||
| Int disable_quality_filtering | |||
| Int pen_cansplice | |||
| Int pen_noncansplice | |||
| Int min_intronlen | |||
| Int max_intronlen | |||
| Int maxins | |||
| Int minins | |||
| Int fastqc_disk_size | |||
| Int fastqscreen_disk_size | |||
| Int qualimapBAMqc_disk_size | |||
| Int qualimapRNAseq_disk_size | |||
| Int insert_size | |||
| Int minimum_length_allowed_for_the_predicted_transcripts | |||
| Int Junctions_no_spliced_reads | |||
| Float minimum_isoform_abundance | |||
| Float maximum_fraction_of_muliplelocationmapped_reads | |||
| String ballgown_docker | |||
| String ballgown_cluster | |||
| String disk_size | |||
| call fastp.fastp as fastp { | |||
| input: | |||
| sample_id=sample_id, | |||
| read1 = read1, | |||
| read2 = read2, | |||
| docker = fastp_docker, | |||
| cluster = fastp_cluster, | |||
| disk_size = disk_size, | |||
| adapter_sequence = adapter_sequence, | |||
| adapter_sequence_r2 = adapter_sequence_r2, | |||
| umi_loc = umi_loc, | |||
| trim_front1 = trim_front1, | |||
| trim_tail1 = trim_tail1, | |||
| max_len1 = max_len1, | |||
| trim_front2 = trim_front2, | |||
| trim_tail2 = trim_tail2, | |||
| max_len2 = max_len2, | |||
| disable_adapter_trimming = disable_adapter_trimming, | |||
| length_required = length_required, | |||
| umi_len = umi_len, | |||
| UMI = UMI, | |||
| qualified_quality_phred = qualified_quality_phred, | |||
| length_required1 = length_required1, | |||
| disable_quality_filtering = disable_quality_filtering | |||
| } | |||
| call fastqc.fastqc as fastqc { | |||
| input: | |||
| read1 = fastp.Trim_R1, | |||
| read2 = fastp.Trim_R2, | |||
| docker = fastqc_docker, | |||
| cluster_config = fastqc_cluster_config, | |||
| disk_size = fastqc_disk_size | |||
| } | |||
| call fastqscreen.fastq_screen as fastqscreen { | |||
| input: | |||
| read1 = fastp.Trim_R1, | |||
| read2 = fastp.Trim_R2, | |||
| screen_ref_dir = screen_ref_dir, | |||
| fastq_screen_conf = fastq_screen_conf, | |||
| docker = fastqscreen_docker, | |||
| cluster_config = fastqscreen_cluster_config, | |||
| disk_size = fastqscreen_disk_size | |||
| } | |||
| call hisat2.hisat2 as hisat2 { | |||
| input: | |||
| sample_id = sample_id, | |||
| idx = idx, | |||
| idx_prefix = idx_prefix, | |||
| Trim_R1 = fastp.Trim_R1, | |||
| Trim_R2 = fastp.Trim_R2, | |||
| docker = hisat2_docker, | |||
| cluster = hisat2_cluster, | |||
| disk_size = disk_size, | |||
| pen_intronlen = pen_intronlen, | |||
| pen_cansplice = pen_cansplice, | |||
| pen_noncansplice = pen_noncansplice, | |||
| min_intronlen = min_intronlen, | |||
| max_intronlen = max_intronlen, | |||
| maxins = maxins, | |||
| minins = minins | |||
| } | |||
| call samtools.samtools as samtools { | |||
| input: | |||
| sample_id = sample_id, | |||
| sam = hisat2.sam, | |||
| docker = samtools_docker, | |||
| cluster = samtools_cluster, | |||
| disk_size = disk_size, | |||
| insert_size = insert_size | |||
| } | |||
| call qualimapBAMqc.qualimapBAMqc as qualimapBAMqc { | |||
| input: | |||
| bam = samtools.out_percent, | |||
| docker = qualimapBAMqc_docker, | |||
| cluster_config = qualimapBAMqc_cluster_config, | |||
| disk_size = qualimapBAMqc_disk_size | |||
| } | |||
| call qualimapRNAseq.qualimapRNAseq as qualimapRNAseq { | |||
| input: | |||
| bam = samtools.out_percent, | |||
| docker = qualimapRNAseq_docker, | |||
| cluster_config = qualimapRNAseq_cluster_config, | |||
| disk_size = qualimapRNAseq_disk_size, | |||
| gtf = gtf | |||
| } | |||
| call stringtie.stringtie as stringtie { | |||
| input: | |||
| sample_id = sample_id, | |||
| gtf = gtf, | |||
| bam = samtools.out_bam, | |||
| docker = stringtie_docker, | |||
| cluster = stringtie_cluster, | |||
| disk_size = disk_size, | |||
| minimum_length_allowed_for_the_predicted_transcripts = minimum_length_allowed_for_the_predicted_transcripts, | |||
| Junctions_no_spliced_reads = Junctions_no_spliced_reads, | |||
| minimum_isoform_abundance = minimum_isoform_abundance, | |||
| maximum_fraction_of_muliplelocationmapped_reads = maximum_fraction_of_muliplelocationmapped_reads | |||
| } | |||
| call ballgown.ballgown as ballgown { | |||
| input: | |||
| sample_id = sample_id, | |||
| docker = ballgown_docker, | |||
| cluster = ballgown_cluster, | |||
| ballgown = stringtie.ballgown, | |||
| gene_abundance = stringtie.gene_abundance, | |||
| disk_size = disk_size | |||
| } | |||
| } | |||
Loading…
